


生物技术进展 ›› 2022, Vol. 12 ›› Issue (6): 806-816.DOI: 10.19586/j.2095-2341.2022.0056
收稿日期:2022-04-14
接受日期:2022-08-30
出版日期:2022-11-25
发布日期:2022-11-30
通讯作者:
朱建华
作者简介:杨洋 E-mail: yang3317@qq.com;
基金资助:
Yang YANG( ), Fenglin WANG, De LIU, Yuanyuan LUO, Jianhua ZHU(
), Fenglin WANG, De LIU, Yuanyuan LUO, Jianhua ZHU( )
)
Received:2022-04-14
Accepted:2022-08-30
Online:2022-11-25
Published:2022-11-30
Contact:
Jianhua ZHU
摘要:
基因编辑(gene editing)技术可以对目的基因进行定点插入、敲除和置换。基于CRISPR-Cas9的基因编辑技术是继锌指核酸酶和转录激活样效应物核酸酶之后的第3代基因编辑技术。近年来,CRISPR-Cas9系统作为研究的热点被广泛应用于医学、药学、植物学、动物学和微生物学等领域,但其在植物次生代谢物领域的应用还处于探索时期。阐述了基于CRISPR-Cas9基因编辑技术的发展历程、工作原理和几种常用的基因编辑方法及其应用实例,总结了CRISPR-Cas9技术在对植物次生代谢产物研究方面的应用。利用CRISPR-Cas9系统可对植物基因组进行定点敲除、突变和插入,以达到提高植物次生代谢物含量、改良作物品质和提高植物抗性等目的。该技术已在植物次生代谢物生物合成关键酶基因的编辑等方面显示出越来越重要的作用。
中图分类号:
杨洋, 王凤林, 刘德, 罗园园, 朱建华. CRISPR⁃Cas9技术在植物次生代谢物生产中的研究进展[J]. 生物技术进展, 2022, 12(6): 806-816.
Yang YANG, Fenglin WANG, De LIU, Yuanyuan LUO, Jianhua ZHU. Research Progress of CRISPR⁃Cas9 Technology on the Production of Plant Secondary Metabolites[J]. Current Biotechnology, 2022, 12(6): 806-816.
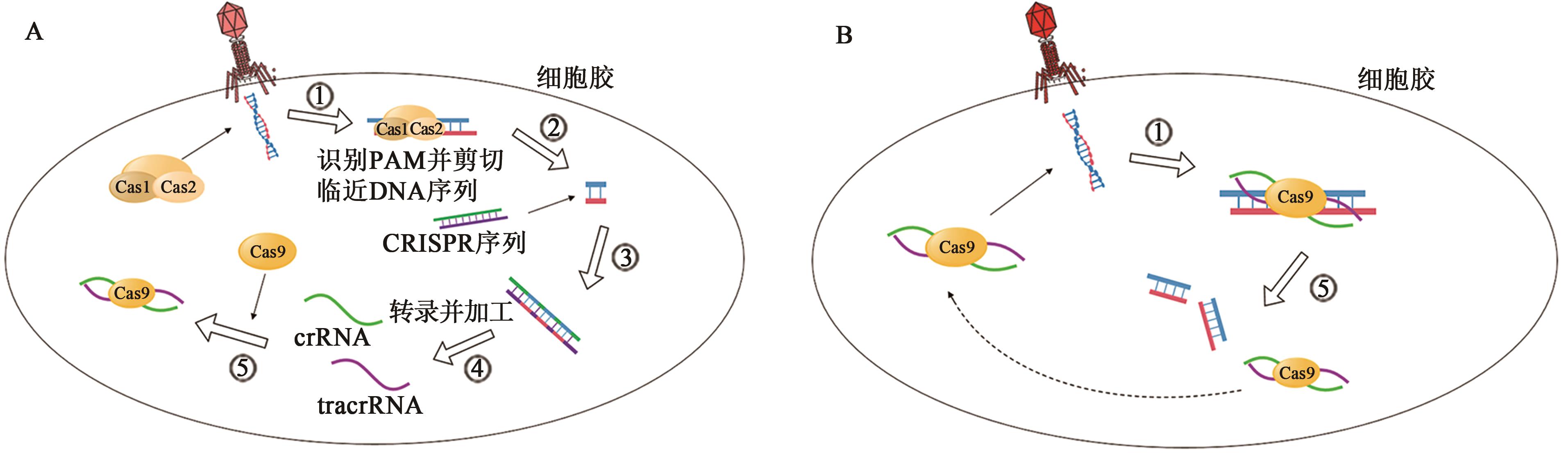
图1 天然CRISPR?Cas9系统[19]A:外源DNA首次入侵细菌,产生crRNA/tracrRNA/Cas9复合物。①—Cas1和Cas2蛋白识别外源DNA的PAM序列;②—识别后剪切DNA临近序列;③—在其他酶的协助下将剪切下来的DNA片段作为间隔序列插入到临近的CRISPR序列中,然后进行DNA修复,修补双链缺口;④—细菌自身附近的CRISPR序列经转录和加工表达crRNA和tracrRNA;⑤—crRNA、tracrRNA和Cas9蛋白组合成为复合物备用。B:相同外源DNA再次入侵细菌后靶基因的识别与切割。①—crRNA/tracrRNA/Cas9复合物识别并精确定位至与crRNA互补的外源DNA的PAM序列;②—crRNA/tracrRNA/Cas9复合物对外源DNA进行精确切割,使外源DNA双链断裂而失活。
Fig. 1 Natural CRISPR?Cas9 system[19]
| 1 | WOODFORD N, ELLINGTON M J. The emergence of antibiotic resistance by mutation[J]. Clin. Microbiol. Infect., 2007,13(1): 5-18. |
| 2 | BAK R O, GOMEZ-OSPINA N, PORTEUS M H. Gene editing on center stage[J]. Trends Genet., 2018, 34(8): 600-611. |
| 3 | STODDARD B L. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification[J]. Structure, 2011,19(1): 7-15. |
| 4 | KIM S, LEE M J, KIM H, et al.. Preassembled zinc-finger arrays for rapid construction of ZFNs[J/OL]. Nat. Methods, 2011, 8(1): 7 [2022-01-18]. . |
| 5 | BOCH J, SCHOLZE H, SCHORNACK S, et al.. Breaking the code of DNA binding specificity of TAL-type Ⅲ effectors[J]. Science, 2009, 326(5959): 1509-1512. |
| 6 | JINEK M, CHYLINSKI K, FONFARA I, et al.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 7 | CARROLL D. Genome engineering with targetable nucleases[J]. Annu. Rev. Biochem., 2014, 83(1): 409-439. |
| 8 | GAJ T, GERSBACH C A, BARBAS C F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering[J]. Trends Biotechnol., 2013, 31(7): 397-405. |
| 9 | 杜丽娜,张存莉,朱玮,等.植物次生代谢合成途径及生物学意义[J].西北林学院学报, 2005(3): 150-155. |
| 10 | 孙立影,于志晶,李海云,等.植物次生代谢物研究进展[J].吉林农业科学, 2009, 34(4): 4-10. |
| 11 | LI J F, NORVILLE J E, AACH J, et al.. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9[J]. Nat. Biotechnol., 2013, 31(8): 688-691. |
| 12 | 胡黎明,彭彦,郑文杰,等. 利用CRISPR/Cas9技术创制镉低积累香型水稻[J]. 杂交水稻,2021,36(6):77-83. |
| 13 | 李红英,高延武,于茹恩,等.利用CRISPR/Cas9技术创建拟南芥Argonaute2基因缺失突变体[J].浙江农业学报, 2021, 33(11): 2001-2008. |
| 14 | 冯吉,程玲,蔡长春,等. 基于CRISPR/Cas9技术的烟草烟碱相关基因敲除及功能研究[J]. 中国烟草科学, 2021,42(2): 84-90. |
| 15 | 曹巧,史占良,张国丛,等. CRISPR/Cas9技术在小麦育种中的应用进展[J].生物技术进展, 2021, 11(6): 661-667. |
| 16 | CAI Y, CHEN L, SUN S, et al.. CRISPR/Cas9-mediated deletion of large genomic fragments in soybean[J]. Int. J. Mol. Sci., 2018, 19(12): 3835. |
| 17 | 娄红梅,杨庆玲,向小雪.番茄MYB44基因敲除载体构建研究[J].特种经济动植物, 2022, 25(1): 19-22. |
| 18 | 王莹婕,马玲玲,梁振.CRISPR/Cas9基因组编辑技术及其在作物遗传改良中的应用进展[J].山西农业科学, 2021, 49(12): 1383-1392. |
| 19 | ISHINO Y, SHINAGAWA H, MAKINO K, et al.. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. J. Bacteriol., 1987, 169(12): 5429-5433. |
| 20 | VAN BELKUM A, SCHERER S, VAN ALPHEN L, et al.. Short-sequence DNA repeats in prokaryotic genomes[J]. Microbiol. Mol. Biol. Rev., 1998, 62(2): 275-293. |
| 21 | JANSEN R, EMBDEN J D, GAASTRA W, et al.. Identification of genes that are associated with DNA repeats in prokaryotes[J]. Mol. Microbiol., 2002, 43(6): 1565-1575. |
| 22 | ZHANG S, GUO F, YAN W, et al.. Recent advances of CRISPR/Cas9-based genetic engineering and transcriptional regulation in industrial biology[J/OL]. Front. Bioeng. Biotechnol., 2020,7(1): 459 [2022-01-18]. . |
| 23 | JIANG F, DOUDNA J A. CRISPR-Cas9 structures and mechanisms[J]. Annu. Rev. Biophys., 2017, 46(1): 505-529. |
| 24 | LOMBARDI L, TURNER S A, ZHAO F, et al.. Gene editing in clinical isolates of Candida parapsilosis using CRISPR/Cas9[J/OL]. Sci. Rep., 2017, 7(1): 8051[2022-01-18]. . |
| 25 | VYAS V K, BUSHKIN G G, BERNSTEIN D A, et al.. New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi[J]. Msphere, 2018, 3(2): e00154-18. |
| 26 | SYMINGTON L S, GAUTIER J. Double-strand break end resection and repair pathway choice[J]. Annu. Rev. Genet., 2011, 45(1): 247-271. |
| 27 | 方锐,畅飞,孙照霖,等.CRISPR/Cas9介导的基因组定点编辑技术[J].生物化学与生物物理进展, 2013, 40(8): 691-702. |
| 28 | 马笃军,彭力平,周紫琼,等.基于CRISPR/Cas9技术编辑RAB39B基因对骨髓间充质干细胞软骨分化的影响[J].中国组织工程研究, 2022, 26(19): 2978-2984. |
| 29 | MIAO C, XIAO L, HUA K, et al.. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity[J]. Proc. Natl. Acad. Sci. USA, 2018, 115(23): 6058-6063. |
| 30 | ZHAO H, LI Y, HE L, et al.. In vivo AAV-CRISPR/Cas9-mediated gene editing Ameliorates atherosclerosis in familial hypercholesterolemia[J]. Circulation, 2020, 41(1): 67-79. |
| 31 | KOMOR A C, KIM Y B, PACKER M S, et al.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| 32 | PORTO E M, KOMOR A C, SLAYMAKER I M, et al.. Base editing: advances and therapeutic opportunities[J]. Nat. Rev. Drug Discov., 2020, 19(12): 839-859. |
| 33 | YARRA R, SAHOO L. Base editing in rice: current progress, advances, limitations, and future perspectives[J]. Plant Cell Rep., 2021, 40(4): 595-604. |
| 34 | WANG Y, LIU Y, ZHENG P, et al.. Microbial base editing: a powerful emerging technology formicrobial genome engineering[J]. Trends Biotechnol., 2021, 39(2): 165-180. |
| 35 | MAKAROVA K S, HAFT D H, BARRANGOU R, et al.. Evolution and classification of the CRISPR-Cas systems[J]. Nat. Rev. Microbiol., 2011, 9(6): 467-77. |
| 36 | ČERMÁK T, BALTES N J, ČEGAN R, et al.. High-frequency, precise modification of the tomato genome[J]. Genome Biol., 2015, 16(11): 1-15. |
| 37 | YU Z, CHEN Q, CHEN W, et al.. Multigene editing via CRISPR/Cas9 guided by a single-sgRNA seed in Arabidopsis [J]. J. Integr. Plant Biol., 2018, 60(5): 376-381. |
| 38 | 陈稷,张浪,黄进.兰科植物转基因技术研究进展[J].生物学教学, 2022, 47(4): 2-5. |
| 39 | 邹俊杰,徐妙云,张兰,等.转基因抗虫、耐除草剂及品质改良复合性状玉米BBHTL8-1的分子特征及功能评价[J].中国农业科技导报, 2022, 24(2): 77-85. |
| 40 | 叶兴国,杜丽璞.从转基因技术角度谈转基因植物的安全性[J].中国种业, 2016(9): 12-13. |
| 41 | LI L N. Biologically active components from traditional Chinese medicines[J]. Pure Appl. Chem., 2013, 70(3): 547-554. |
| 42 | GAO W, HILLWIG M L, HUANG L Q, et al.. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights[J]. Org. Lett., 2009, 11(22): 5170-5173. |
| 43 | LI B, CUI G, SHEN G, et al.. Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza [J]. Sci. Rep., 2017, 7(1): 43320. |
| 44 | LI B, LI J, CHAI Y, HUANG Y, et al.. Targeted mutagenesis of CYP76AK2 and CYP76AK3 in Salvia miltiorrhiza reveals their roles in tanshinones biosynthetic pathway[J]. Int. J. Biol. Macromol., 2021, 189(1): 455-463. |
| 45 | 代卓毅,李洪亮,姚怡帆,等.编辑烤烟多酚氧化酶基因NtPPO8后的效应分析[J].中国烟草学报, 2022, 28(5): 73-83. |
| 46 | ADMAN E T, STENKAMP R E, SIEKER L C, et al.. A crystallographic model for azurin a 3A resolution[J]. J. Mol. Biol., 1978, 123(1): 35-47. |
| 47 | DENG C, SHI M, FU R, et al.. ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza [J]. J. Exp. Bot., 2020, 71(19): 5948-5962. |
| 48 | ZHOU Z, TAN H, LI Q, et al.. CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza [J]. Phytochemistry, 2018, 148(1): 63-70. |
| 49 | ZENG L, ZHANG Q, JIANG C, et al.. Development of Atropa belladonna L. Plants with high-yield hyoscyamine and without its derivatives using the CRISPR/Cas9 system[J/OL]. Int. J. Mol. Sci., 2021, 22(4): 1731[2021-02-09]. . |
| 50 | MA W, KANG X, LIU P, et al.. The analysis of transcription factor CsHB1 effects on caffeine accumulation in tea callus through CRISPR/Cas9 mediated gene editing[J]. Proc. Biochem., 2021, 101(1): 304-311. |
| 51 | STAIGER C. Comfrey: a clinical overview[J]. Phytother. Res., 2012, 26(10): 1441-1448. |
| 52 | AVILA C, BREAKSPEAR I, HAWRELAK J, et al.. A systematic review and quality assessment of case reports of adverse events for borage (Borago officinalis), coltsfoot (Tussilago farfara) and comfrey (Symphytum officinale)[J/OL]. Fitoterapia, 2020, 142:104519[2020-02-24]. . |
| 53 | ZAKARIA M M, SCHEMMERLING B, OBER D, et al.. CRISPR/Cas9-mediated genome editing in comfrey (Symphytum officinale) hairy roots results in the complete eradication of pyrrolizidine alkaloids[J/OL]. Molecules, 2021, 26(6): 1498[2021-03-10]. . |
| 54 | KOPP T, ABDEL-TAWAB M, MIZAIKOFF B. Extracting and analyzing pyrrolizidine alkaloids in medicinal plants: a review[J]. Toxins (Basel), 2020, 12(5): 320. |
| 55 | 王威威,席飞虎,杨少峰,等. 烟草烟碱合成代谢调控研究进展[J]. 亚热带农业研究, 2016, 12(1): 62-67. |
| 56 | CHEN S, YANG Y, SHI W, et al.. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance[J]. Plant Cell, 2008, 20(7): 1850-1861. |
| 57 | MAHATTANATAWEE K, ROUSEFF R L. Comparison of aroma active and sulfur volatiles in three fragrant rice cultivars using GC-olfactometry and GC-PFPD[J]. Food Chem., 2014, 154(1): 1-6. |
| 58 | 张翔,史亚兴,卢柏山,等.利用CRISPR/Cas9技术编辑BADH2-1/BADH2-2创制香米味道玉米新种质[J].中国农业科学, 2021, 54(10): 2064-2075. |
| 59 | TU Y. Artemisinin- a gift from traditional Chinese medicine to the world (nobel. ecture)[J]. Angew. Chem. Int. Ed. Engl., 2016, 55(35): 10210-10226. |
| 60 | WANG J G, XU C C, WONG Y K, et al.. Artemisinin, the magic drug discovered from traditional Chinese medicine[J]. Engineering, 2019, 5(1): 32-39. |
| 61 | SONG Y F, HE S Q, Abdallah I I, et al.. Engineering of multiple modules to improve amorphadiene production in Bacillus subtilis using CRISPR-Cas9[J]. J. Agric. Food Chem., 2021, 69(16): 4785-4794. |
| 62 | 杨林,汪逗逗,田少凯,等. 甘草DXS基因过表达及表达沉默对甘草酸生物合成的影响研究[J]. 药学学报, 2021, 56(7): 2025-2032. |
| 63 | LIU Y, ZHOU J, HU T Y. Identification and functional characterization of squalene epoxidases and oxidosqualene cyclases from Tripterygium wilfordii [J]. Plant Cell Rep., 2020, 39(3): 409-418. |
| 64 | ROBERTS S C. Production and engineering of terpenoids in plant cell culture[J]. Nat. Chem. Biol., 2007, 3(7): 387-395. |
| 65 | 张冠华,刁倩楠. 类胡萝卜素研究进展[J]. 现代农业, 2021(4): 46-49. |
| 66 | WATANABE K, ODA-YAMAMIZO C, SAGE-ONO K, et al.. Alteration of flower colour in Ipomoea nil through CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 4[J]. Transgenic. Res., 2018, 27(1): 25-38. |
| 67 | 左鑫,李铭铭,李欣容,等. CRISPR/Cas9技术在天目地黄RcPDS1基因编辑中的应用[J].园艺学报, 2022, 49(7): 1532-1544. |
| 68 | JAYARAJ K L, THULASIDHARAN N, ANTONY A, et al.. Targeted editing of tomato carotenoid isomerase reveals the role of 5′UTR region in gene expression regulation[J]. Plant Cell Rep., 2021, 40(4): 621-635. |
| 69 | MIYAMOTO T, TAKADA R, TOBIMATSU Y, et al.. OsMYB108 loss-of-function enriches p-coumaroylated and tricin lignin units in rice cell walls[J]. Plant J., 2019, 98(6): 975-987. |
| 70 | KUI L, CHEN H, ZHANG W, et al.. Building a genetic manipulation tool bbox for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, dendrobium officinale[J/OL]. Front. Plant Sci., 2017, 7(1): 2036 [2017-01-12]. . |
| 71 | 王琪,张浩,雷秀娟,等. 人参CAS基因CRISPR/Cas9载体的构建及初步转化[J].分子植物育种, 2021,19(8): 2603-2608. |
| 72 | 王琪.应用CRISPR/Cas9技术构建人参CAS基因和三七DS基因的载体及转化体系建立[D].北京:中国农业科学院, 2020. |
| 73 | CHOI H S, KOO H B, JEON S W, et al.. Modification of ginsenoside saponin composition via the CRISPR/Cas9-mediated knockout of protopanaxadiol 6-hydroxylase gene in Panax ginseng [J]. J. Ginseng. Res., 2022, 46(4): 505-514. |
| 74 | FENG S, SONG W, FUR, et al.. Application of the CRISPR/Cas9 system in Dioscorea zingiberensis [J]. Plant Cell Tiss. Org., 2018, 135(1): 133-141. |
| 75 | RIE B J, DANIELE S V, JØRN S, et al.. LC-MS/MS with atmospheric pressure chemical ionisation to study the effect of UV treatment on the formation of vitamin D3 and sterols in plants[J]. Food Chem., 2011, 129(1): 129: 217-225. |
| 76 | LI J, SCARANO A, GONZALEZ N M, et al.. Biofortified tomatoes provide a new route to vitamin D sufficiency[J]. Nat. Plants, 2022, 8(6): 611-616. |
| 77 | YANG X, WANG J, XIA X, et al.. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice[J]. Plant J., 2021, 107(1): 198-214. |
| 78 | 杨迎霞,葛贤宏,孙德岭,等.花椰菜高效CRISPR/Cas9基因编辑技术体系的构建[J].天津农业科学, 2022, 28(4): 5-9. |
| 79 | WEN D, WU L, WANG M, et al.. CRISPR/Cas9-mediated targeted mutagenesis of FtMYB45 promotes flavonoid biosynthesis in tartary buckwheat (Fagopyrum tataricum)[J/OL]. Front. Plant Sci., 2022, 13: 879390[2022-05-12]. . |
| 80 | ZHANG P P, DU H Y, WANG J, et al.. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus[J]. Plant Biotechnol. J., 2020, 18(6): 1384-1395. |
| 81 | DINKINS R D, HANCOK J, COE B L, et al.. Isoflavone levels, nodulation and gene expression profiles of a CRISPR/Cas9 deletion mutant in the isoflavone synthase gene of red clover[J]. Plant Cell Rep., 2021, 40(3): 517-528. |
| [1] | 孙卉, 张春义, 姜凌. 药用植物分子农场研究进展[J]. 生物技术进展, 2023, 13(1): 65-71. |
| [2] | 于鲲, 薛佳琪, 王进宽, 余永涛. CRISPR/Cas9基因编辑技术在丝状真菌中的应用[J]. 生物技术进展, 2022, 12(5): 696-704. |
| [3] | 高维崧, 窦金萍, 韦双, 刘兴健, 张志芳, 李轶女. CRISPR/Cas系统的分类及研究现状[J]. 生物技术进展, 2022, 12(4): 532-538. |
| [4] | 党星, 郅斌伟, 曹克浩, 刘婷婷, 陈飚, 丁远杰. 转基因玉米生物育种技术的专利分析及产业发展建议[J]. 生物技术进展, 2022, 12(4): 614-622. |
| [5] | 黄耀辉, 王艺洁, 杨立桃, 焦悦, 付仲文. 生物育种新技术作物的安全管理[J]. 生物技术进展, 2022, 12(2): 198-204. |
| [6] | 曹巧, 史占良, 张国丛, 班进福, 郑树松, 傅晓艺, 张士昌, 何明琦, 韩然, 高振贤. CRISPR/Cas9技术在小麦育种中的应用进展[J]. 生物技术进展, 2021, 11(6): 661-667. |
| [7] | 林敏. 农业生物育种技术的发展历程及产业化对策[J]. 生物技术进展, 2021, 11(4): 405-417. |
| [8] | 王梦雨, 王颢潜, 王旭静, 王志兴. 基因编辑产品检测技术研究进展[J]. 生物技术进展, 2021, 11(4): 438-445. |
| [9] | 格日乐其木格, 牛振峰, 董丹, 张涛涛, 峥嵘. CRISPR-Cas系统在微生物研究中的应用进展[J]. 生物技术进展, 2021, 11(3): 253-259. |
| [10] | 曹豪豪,张红兵,薛溪发,李左群,闫洪波,李会宣. 新型基因编辑技术在单细胞微藻中的应用进展[J]. 生物技术进展, 2021, 11(1): 9-15. |
| [11] | 薛琴琴,,韩贝贝,吴雪晴,李沛,李莹莹,,吴庆钰. 纳米材料在农作物领域的应用及展望[J]. 生物技术进展, 2020, 10(6): 655-660. |
| [12] | 解美霞,,王燕,赵新,高越,刘双,陈锐,兰青阔,徐晓辉4,曹际娟5,王永. 基于焦磷酸测序技术建立基因编辑水稻检测方法[J]. 生物技术进展, 2020, 10(6): 668-673. |
| [13] | 朱瑶瑶,张朔,王征宇. 利用人胚胎干细胞来源的Runx1c-mNeonGreen报告细胞系追踪成年造血过程[J]. 生物技术进展, 2020, 10(5): 524-533. |
| [14] | 李凯,,罗云波,,许文涛,. CRISPR-Cas生物传感器研究进展[J]. 生物技术进展, 2019, 9(6): 579-591. |
| [15] | 郑红艳,王磊. CRISPR/Cas基因编辑技术及其在作物育种中的应用[J]. 生物技术进展, 2018, 8(3): 185-190. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部