


生物技术进展 ›› 2023, Vol. 13 ›› Issue (3): 412-424.DOI: 10.19586/j.2095-2341.2023.0007
胡文冉1( ), 郝晓燕1, 赵准1, 邵武奎2, 高升旗1, 李建平1, 黄全生1(
), 郝晓燕1, 赵准1, 邵武奎2, 高升旗1, 李建平1, 黄全生1( )
)
收稿日期:2023-02-02
接受日期:2023-02-28
出版日期:2023-05-25
发布日期:2023-06-12
通讯作者:
黄全生
作者简介:胡文冉 E-mail: huwran@126.com;
基金资助:
Wenran HU1( ), Xiaoyan HAO1, Zhun ZHAO1, Wukui SHAO2, Shengqi GAO1, Jianping LI1, Quansheng HUANG1(
), Xiaoyan HAO1, Zhun ZHAO1, Wukui SHAO2, Shengqi GAO1, Jianping LI1, Quansheng HUANG1( )
)
Received:2023-02-02
Accepted:2023-02-28
Online:2023-05-25
Published:2023-06-12
Contact:
Quansheng HUANG
摘要:
苯丙烷代谢是植物重要的次生代谢途径之一,其代谢产物在植物的生长发育中发挥着重要作用。肉桂酸脱氢酶(cinnamyl alcohol dehydrogenase, CAD)是苯丙烷代谢途径的关键酶之一,在棉花纤维品质形成中起着十分重要的调节作用。为了更好地了解CAD基因家族在二倍体雷蒙德氏棉(D5)和亚洲棉(A2)、四倍体陆地棉(AD1)和海岛棉(AD2)基因组中的数量和分布情况,通过生物信息学方法,在雷蒙德氏棉、亚洲棉、陆地棉和海岛棉全基因组中分别鉴定出16、16、28和25个CAD基因家族成员,进一步分析了这些CAD家族成员进行基因结构、染色体定位、保守域、进化关系及CAD基因在陆地棉不同器官组织中的表达等。结果表明,4个棉种全基因组分别编码16、16、28和25个CAD基因。亚细胞定位将这些棉花CAD基因的表达产物均定位于细胞质。雷蒙德氏棉有14个CAD成员基因分布在5条染色体上,亚洲棉有13个CAD成员基因分布在5条染色体上,陆地棉28个CAD成员基因、海岛棉25个CAD基因分别分布在10条染色体上;内含子/外显子结构分析表明,CAD基因结构较为复杂,均含有内含子和外显子;功能结构域分析发现,有棉花CAD蛋白具有高度保守的ADH_N和ADH_zinc_N 2个功能域;根据系统发育分析结果,将CAD基因家族分成3个亚类。大部分CAD基因在陆地棉的不同器官中均有表达,部分在棉花纤维中表达量较高。分析结果为进一步研究棉花CAD基因家族各成员的功能奠定了理论基础。
中图分类号:
胡文冉, 郝晓燕, 赵准, 邵武奎, 高升旗, 李建平, 黄全生. 棉花CAD基因家族的生物信息学分析[J]. 生物技术进展, 2023, 13(3): 412-424.
Wenran HU, Xiaoyan HAO, Zhun ZHAO, Wukui SHAO, Shengqi GAO, Jianping LI, Quansheng HUANG. Bioinformatic Analysis of the CAD Gene Family in Cotton[J]. Current Biotechnology, 2023, 13(3): 412-424.
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GrCAD1 | Cotton_D_gene_10016775 | Chr08 | 1 068 | 355 | 38.45 | 6.65 | 细胞质 |
| GrCAD2 | Cotton_D_gene_10037629 | Chr08 | 942 | 313 | 33.98 | 6.31 | 细胞质 |
| GrCAD3 | Cotton_D_gene_10012650 | Chr07 | 1 095 | 364 | 39.12 | 5.85 | 细胞质 |
| GrCAD4 | Cotton_D_gene_10038273 | Chr08 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GrCAD5 | Cotton_D_gene_10016445 | Chr10 | 1 077 | 358 | 38.70 | 5.64 | 细胞质 |
| GrCAD6 | Cotton_D_gene_10033972 | Chr09 | 1 080 | 359 | 39.19 | 7.13 | 细胞质 |
| GrCAD7 | Cotton_D_gene_10036902 | Chr07 | 1 059 | 352 | 38.18 | 6.12 | 细胞质 |
| GrCAD8 | Cotton_D_gene_10014351 | Chr07 | 780 | 259 | 28.15 | 5.99 | 细胞质 |
| GrCAD9 | Cotton_D_gene_10036599 | Chr03 | 1 080 | 359 | 39.09 | 5.58 | 细胞质 |
| GrCAD10 | Cotton_D_gene_10002391 | scaffold369 | 990 | 329 | 35.61 | 6.07 | 细胞质 |
| GrCAD11 | Cotton_D_gene_10002389 | scaffold369 | 1 086 | 361 | 39.29 | 6.26 | 细胞质 |
| GrCAD12 | Cotton_D_gene_10037630 | Chr08 | 741 | 246 | 27.09 | 8.94 | 细胞质 |
| GrCAD13 | Cotton_D_gene_10037632 | Chr08 | 774 | 257 | 28.02 | 8.12 | 细胞质 |
| GrCAD14 | Cotton_D_gene_10037631 | Chr08 | 1 089 | 362 | 39.10 | 6.50 | 细胞质 |
| GrCAD15 | Cotton_D_gene_10014346 | Chr07 | 882 | 293 | 31.46 | 6.25 | 细胞质 |
| GrCAD16 | Cotton_D_gene_10014349 | Chr07 | 1 095 | 364 | 39.11 | 5.75 | 细胞质 |
表1 雷蒙德氏棉CAD基因的序列特征
Table 1 The characteristic of CAD genes in Gossypium raimondii
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GrCAD1 | Cotton_D_gene_10016775 | Chr08 | 1 068 | 355 | 38.45 | 6.65 | 细胞质 |
| GrCAD2 | Cotton_D_gene_10037629 | Chr08 | 942 | 313 | 33.98 | 6.31 | 细胞质 |
| GrCAD3 | Cotton_D_gene_10012650 | Chr07 | 1 095 | 364 | 39.12 | 5.85 | 细胞质 |
| GrCAD4 | Cotton_D_gene_10038273 | Chr08 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GrCAD5 | Cotton_D_gene_10016445 | Chr10 | 1 077 | 358 | 38.70 | 5.64 | 细胞质 |
| GrCAD6 | Cotton_D_gene_10033972 | Chr09 | 1 080 | 359 | 39.19 | 7.13 | 细胞质 |
| GrCAD7 | Cotton_D_gene_10036902 | Chr07 | 1 059 | 352 | 38.18 | 6.12 | 细胞质 |
| GrCAD8 | Cotton_D_gene_10014351 | Chr07 | 780 | 259 | 28.15 | 5.99 | 细胞质 |
| GrCAD9 | Cotton_D_gene_10036599 | Chr03 | 1 080 | 359 | 39.09 | 5.58 | 细胞质 |
| GrCAD10 | Cotton_D_gene_10002391 | scaffold369 | 990 | 329 | 35.61 | 6.07 | 细胞质 |
| GrCAD11 | Cotton_D_gene_10002389 | scaffold369 | 1 086 | 361 | 39.29 | 6.26 | 细胞质 |
| GrCAD12 | Cotton_D_gene_10037630 | Chr08 | 741 | 246 | 27.09 | 8.94 | 细胞质 |
| GrCAD13 | Cotton_D_gene_10037632 | Chr08 | 774 | 257 | 28.02 | 8.12 | 细胞质 |
| GrCAD14 | Cotton_D_gene_10037631 | Chr08 | 1 089 | 362 | 39.10 | 6.50 | 细胞质 |
| GrCAD15 | Cotton_D_gene_10014346 | Chr07 | 882 | 293 | 31.46 | 6.25 | 细胞质 |
| GrCAD16 | Cotton_D_gene_10014349 | Chr07 | 1 095 | 364 | 39.11 | 5.75 | 细胞质 |
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GaCAD1 | Ga12G0361 | Chr12 | 1 068 | 355 | 38.41 | 7.05 | 细胞质 |
| GaCAD2 | Ga11G1886 | Chr11 | 987 | 328 | 35.17 | 6.56 | 细胞质 |
| GaCAD3 | Ga12G2139 | Chr12 | 1 098 | 365 | 39.70 | 6.22 | 细胞质 |
| GaCAD4 | Ga14G0143 | tig00000498 | 1 188 | 395 | 42.93 | 5.26 | 细胞质 |
| GaCAD5 | Ga03G2431 | Chr03 | 1 077 | 358 | 38.84 | 5.53 | 细胞质 |
| GaCAD6 | Ga05G1953 | Chr05 | 1 074 | 357 | 39.12 | 6.46 | 细胞质 |
| GaCAD7 | Ga02G0337 | Chr02 | 1 065 | 354 | 39.74 | 6.13 | 细胞质 |
| GaCAD8 | Ga14G1938 | tig00016738 | 339 | 112 | 11.76 | 9.10 | 细胞质 |
| GaCAD9 | Ga02G0499 | Chr02 | 1 080 | 359 | 39.07 | 6.01 | 细胞质 |
| GaCAD10 | Ga02G0339 | Chr02 | 1 086 | 361 | 39.22 | 6.26 | 细胞质 |
| GaCAD11 | Ga14G1937 | tig00016738 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GaCAD12 | Ga12G2141 | Chr12 | 945 | 314 | 34.10 | 6.45 | 细胞质 |
| GaCAD13 | Ga12G2140 | Chr12 | 1 065 | 354 | 37.94 | 7.15 | 细胞质 |
| GaCAD14 | Ga12G2105 | Chr12 | 1 107 | 368 | 39.62 | 6.64 | 细胞质 |
| GaCAD15 | Ga11G1713 | Chr11 | 783 | 260 | 28.16 | 7.17 | 细胞质 |
| GaCAD16 | Ga11G1714 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
表2 亚洲棉CAD基因的序列特征
Table 2 The characteristic of CAD genes in Gossypium arboretum
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GaCAD1 | Ga12G0361 | Chr12 | 1 068 | 355 | 38.41 | 7.05 | 细胞质 |
| GaCAD2 | Ga11G1886 | Chr11 | 987 | 328 | 35.17 | 6.56 | 细胞质 |
| GaCAD3 | Ga12G2139 | Chr12 | 1 098 | 365 | 39.70 | 6.22 | 细胞质 |
| GaCAD4 | Ga14G0143 | tig00000498 | 1 188 | 395 | 42.93 | 5.26 | 细胞质 |
| GaCAD5 | Ga03G2431 | Chr03 | 1 077 | 358 | 38.84 | 5.53 | 细胞质 |
| GaCAD6 | Ga05G1953 | Chr05 | 1 074 | 357 | 39.12 | 6.46 | 细胞质 |
| GaCAD7 | Ga02G0337 | Chr02 | 1 065 | 354 | 39.74 | 6.13 | 细胞质 |
| GaCAD8 | Ga14G1938 | tig00016738 | 339 | 112 | 11.76 | 9.10 | 细胞质 |
| GaCAD9 | Ga02G0499 | Chr02 | 1 080 | 359 | 39.07 | 6.01 | 细胞质 |
| GaCAD10 | Ga02G0339 | Chr02 | 1 086 | 361 | 39.22 | 6.26 | 细胞质 |
| GaCAD11 | Ga14G1937 | tig00016738 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GaCAD12 | Ga12G2141 | Chr12 | 945 | 314 | 34.10 | 6.45 | 细胞质 |
| GaCAD13 | Ga12G2140 | Chr12 | 1 065 | 354 | 37.94 | 7.15 | 细胞质 |
| GaCAD14 | Ga12G2105 | Chr12 | 1 107 | 368 | 39.62 | 6.64 | 细胞质 |
| GaCAD15 | Ga11G1713 | Chr11 | 783 | 260 | 28.16 | 7.17 | 细胞质 |
| GaCAD16 | Ga11G1714 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GhCAD1 | GH_D12G2651 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GhCAD2 | GH_D12G0806 | Chr12 | 945 | 314 | 33.66 | 6.59 | 细胞质 |
| GhCAD3 | GH_A12G0990 | Chr12 | 1 086 | 361 | 39.24 | 6.60 | 细胞质 |
| GhCAD4 | GH_A12G0204 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GhCAD5 | GH_A03G2100 | Chr03 | 1 077 | 358 | 38.81 | 5.53 | 细胞质 |
| GhCAD6 | GH_D05G1877 | Chr05 | 1 080 | 359 | 39.18 | 7.13 | 细胞质 |
| GhCAD7 | GH_D11G2110 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GhCAD8 | GH_D11G2243 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GhCAD9 | GH_D03G0499 | Chr03 | 1 080 | 359 | 39.09 | 5.85 | 细胞质 |
| GhCAD10 | GH_A02G1713 | Chr02 | 1 086 | 361 | 39.28 | 6.00 | 细胞质 |
| GhCAD11 | GH_A11G2195 | Chr11 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GhCAD12 | GH_D12G0807 | Chr12 | 549 | 182 | 19.91 | 7.65 | 细胞质 |
| GhCAD13 | GH_A12G0991 | Chr12 | 1 398 | 465 | 50.54 | 6.02 | 细胞质 |
| GhCAD14 | GH_D12G0808 | Chr12 | 1 089 | 362 | 38.99 | 6.46 | 细胞质 |
| GhCAD15 | GH_A11G2192 | Chr11 | 831 | 276 | 29.86 | 6.59 | 细胞质 |
| GhCAD16 | GH_A11G2193 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GhCAD17 | GH_D03G0349 | Chr03 | 972 | 323 | 34.96 | 6.71 | 细胞质 |
| GhCAD18 | GH_D03G0347 | Chr03 | 1 086 | 361 | 39.19 | 6.12 | 细胞质 |
| GhCAD19 | GH_D11G3241 | Chr11 | 1 095 | 364 | 39.13 | 5.85 | 细胞质 |
| GhCAD20 | GH_A05G1840 | Chr05 | 1 080 | 359 | 39.05 | 6.41 | 细胞质 |
| GhCAD21 | GH_A12G2629 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GhCAD22 | GH_A11G2344 | Chr11 | 894 | 297 | 31.97 | 6.23 | 细胞质 |
| GhCAD23 | GH_D12G0217 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GhCAD24 | GH_A02G1628 | Chr02 | 1 080 | 359 | 39.07 | 5.71 | 细胞质 |
| GhCAD25 | GH_D02G2271 | Chr02 | 663 | 220 | 24.01 | 5.70 | 细胞质 |
| GhCAD26 | GH_D11G2246 | Chr11 | 747 | 248 | 26.73 | 7.14 | 细胞质 |
| GhCAD27 | GH_A12G0994 | Chr12 | 1 137 | 378 | 40.94 | 6.49 | 细胞质 |
| GhCAD28 | GH_A12G0993 | Chr12 | 1 014 | 337 | 36.49 | 6.08 | 细胞质 |
表3 陆地棉CAD基因的序列特征
Table 3 The characteristic of CAD genes in Gossypium hirsutum
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GhCAD1 | GH_D12G2651 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GhCAD2 | GH_D12G0806 | Chr12 | 945 | 314 | 33.66 | 6.59 | 细胞质 |
| GhCAD3 | GH_A12G0990 | Chr12 | 1 086 | 361 | 39.24 | 6.60 | 细胞质 |
| GhCAD4 | GH_A12G0204 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GhCAD5 | GH_A03G2100 | Chr03 | 1 077 | 358 | 38.81 | 5.53 | 细胞质 |
| GhCAD6 | GH_D05G1877 | Chr05 | 1 080 | 359 | 39.18 | 7.13 | 细胞质 |
| GhCAD7 | GH_D11G2110 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GhCAD8 | GH_D11G2243 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GhCAD9 | GH_D03G0499 | Chr03 | 1 080 | 359 | 39.09 | 5.85 | 细胞质 |
| GhCAD10 | GH_A02G1713 | Chr02 | 1 086 | 361 | 39.28 | 6.00 | 细胞质 |
| GhCAD11 | GH_A11G2195 | Chr11 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GhCAD12 | GH_D12G0807 | Chr12 | 549 | 182 | 19.91 | 7.65 | 细胞质 |
| GhCAD13 | GH_A12G0991 | Chr12 | 1 398 | 465 | 50.54 | 6.02 | 细胞质 |
| GhCAD14 | GH_D12G0808 | Chr12 | 1 089 | 362 | 38.99 | 6.46 | 细胞质 |
| GhCAD15 | GH_A11G2192 | Chr11 | 831 | 276 | 29.86 | 6.59 | 细胞质 |
| GhCAD16 | GH_A11G2193 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GhCAD17 | GH_D03G0349 | Chr03 | 972 | 323 | 34.96 | 6.71 | 细胞质 |
| GhCAD18 | GH_D03G0347 | Chr03 | 1 086 | 361 | 39.19 | 6.12 | 细胞质 |
| GhCAD19 | GH_D11G3241 | Chr11 | 1 095 | 364 | 39.13 | 5.85 | 细胞质 |
| GhCAD20 | GH_A05G1840 | Chr05 | 1 080 | 359 | 39.05 | 6.41 | 细胞质 |
| GhCAD21 | GH_A12G2629 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GhCAD22 | GH_A11G2344 | Chr11 | 894 | 297 | 31.97 | 6.23 | 细胞质 |
| GhCAD23 | GH_D12G0217 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GhCAD24 | GH_A02G1628 | Chr02 | 1 080 | 359 | 39.07 | 5.71 | 细胞质 |
| GhCAD25 | GH_D02G2271 | Chr02 | 663 | 220 | 24.01 | 5.70 | 细胞质 |
| GhCAD26 | GH_D11G2246 | Chr11 | 747 | 248 | 26.73 | 7.14 | 细胞质 |
| GhCAD27 | GH_A12G0994 | Chr12 | 1 137 | 378 | 40.94 | 6.49 | 细胞质 |
| GhCAD28 | GH_A12G0993 | Chr12 | 1 014 | 337 | 36.49 | 6.08 | 细胞质 |
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GbCAD1 | Gbar_D12G025360 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GbCAD2 | Gbar_A02G015780 | Chr02 | 981 | 326 | 36.30 | 6.21 | 细胞质 |
| GbCAD3 | Gbar_A12G009020 | Chr12 | 921 | 306 | 33.01 | 6.25 | 细胞质 |
| GbCAD4 | Gbar_A12G002060 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GbCAD5 | Gbar_A03G020480 | Chr03 | 1 077 | 358 | 38.82 | 5.53 | 细胞质 |
| GbCAD6 | Gbar_D05G018810 | Chr05 | 1 035 | 344 | 37.76 | 6.55 | 细胞质 |
| GbCAD7 | Gbar_D11G021150 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GbCAD8 | Gbar_D11G022440 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GbCAD9 | Gbar_D03G004820 | Chr03 | 984 | 327 | 35.62 | 6.19 | 细胞质 |
| GbCAD10 | Gbar_A02G015760 | Chr02 | 1 086 | 361 | 39.33 | 6.26 | 细胞质 |
| GbCAD11 | Gbar_A11G021420 | Chr11 | 1 056 | 351 | 37.98 | 8.63 | 细胞质 |
| GbCAD12 | Gbar_D12G008930 | Chr12 | 549 | 182 | 19.94 | 7.65 | 细胞质 |
| GbCAD13 | Gbar_A12G009010 | Chr12 | 978 | 325 | 34.74 | 6.64 | 细胞质 |
| GbCAD14 | Gbar_D12G008920 | Chr12 | 1 122 | 373 | 40.12 | 6.23 | 细胞质 |
| GbCAD15 | Gbar_D11G022460 | Chr11 | 408 | 135 | 14.46 | 5.62 | 细胞质 |
| GbCAD16 | Gbar_A11G021400 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GbCAD17 | Gbar_D03G003450 | Chr03 | 792 | 263 | 28.34 | 6.15 | 细胞质 |
| GbCAD18 | Gbar_D03G003430 | Chr03 | 1 086 | 361 | 39.16 | 6.12 | 细胞质 |
| GbCAD19 | Gbar_D11G031180 | Chr11 | 1 098 | 365 | 39.26 | 5.75 | 细胞质 |
| GbCAD20 | Gbar_A05G018170 | Chr05 | 1 080 | 359 | 39.17 | 6.82 | 细胞质 |
| GbCAD21 | Gbar_A12G025440 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GbCAD22 | Gbar_D02G022410 | Chr02 | 1 077 | 358 | 38.82 | 5.64 | 细胞质 |
| GbCAD23 | Gbar_D12G002300 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GbCAD24 | Gbar_A11G022760 | Chr11 | 561 | 186 | 19.77 | 7.99 | 细胞质 |
| GbCAD25 | Gbar_A02G014990 | Chr02 | 996 | 331 | 36.10 | 6.46 | 细胞质 |
表4 海岛棉CAD基因的序列特征
Table 4 The characteristic of CAD genes in Gossypium barbadense
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GbCAD1 | Gbar_D12G025360 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GbCAD2 | Gbar_A02G015780 | Chr02 | 981 | 326 | 36.30 | 6.21 | 细胞质 |
| GbCAD3 | Gbar_A12G009020 | Chr12 | 921 | 306 | 33.01 | 6.25 | 细胞质 |
| GbCAD4 | Gbar_A12G002060 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GbCAD5 | Gbar_A03G020480 | Chr03 | 1 077 | 358 | 38.82 | 5.53 | 细胞质 |
| GbCAD6 | Gbar_D05G018810 | Chr05 | 1 035 | 344 | 37.76 | 6.55 | 细胞质 |
| GbCAD7 | Gbar_D11G021150 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GbCAD8 | Gbar_D11G022440 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GbCAD9 | Gbar_D03G004820 | Chr03 | 984 | 327 | 35.62 | 6.19 | 细胞质 |
| GbCAD10 | Gbar_A02G015760 | Chr02 | 1 086 | 361 | 39.33 | 6.26 | 细胞质 |
| GbCAD11 | Gbar_A11G021420 | Chr11 | 1 056 | 351 | 37.98 | 8.63 | 细胞质 |
| GbCAD12 | Gbar_D12G008930 | Chr12 | 549 | 182 | 19.94 | 7.65 | 细胞质 |
| GbCAD13 | Gbar_A12G009010 | Chr12 | 978 | 325 | 34.74 | 6.64 | 细胞质 |
| GbCAD14 | Gbar_D12G008920 | Chr12 | 1 122 | 373 | 40.12 | 6.23 | 细胞质 |
| GbCAD15 | Gbar_D11G022460 | Chr11 | 408 | 135 | 14.46 | 5.62 | 细胞质 |
| GbCAD16 | Gbar_A11G021400 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GbCAD17 | Gbar_D03G003450 | Chr03 | 792 | 263 | 28.34 | 6.15 | 细胞质 |
| GbCAD18 | Gbar_D03G003430 | Chr03 | 1 086 | 361 | 39.16 | 6.12 | 细胞质 |
| GbCAD19 | Gbar_D11G031180 | Chr11 | 1 098 | 365 | 39.26 | 5.75 | 细胞质 |
| GbCAD20 | Gbar_A05G018170 | Chr05 | 1 080 | 359 | 39.17 | 6.82 | 细胞质 |
| GbCAD21 | Gbar_A12G025440 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GbCAD22 | Gbar_D02G022410 | Chr02 | 1 077 | 358 | 38.82 | 5.64 | 细胞质 |
| GbCAD23 | Gbar_D12G002300 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GbCAD24 | Gbar_A11G022760 | Chr11 | 561 | 186 | 19.77 | 7.99 | 细胞质 |
| GbCAD25 | Gbar_A02G014990 | Chr02 | 996 | 331 | 36.10 | 6.46 | 细胞质 |
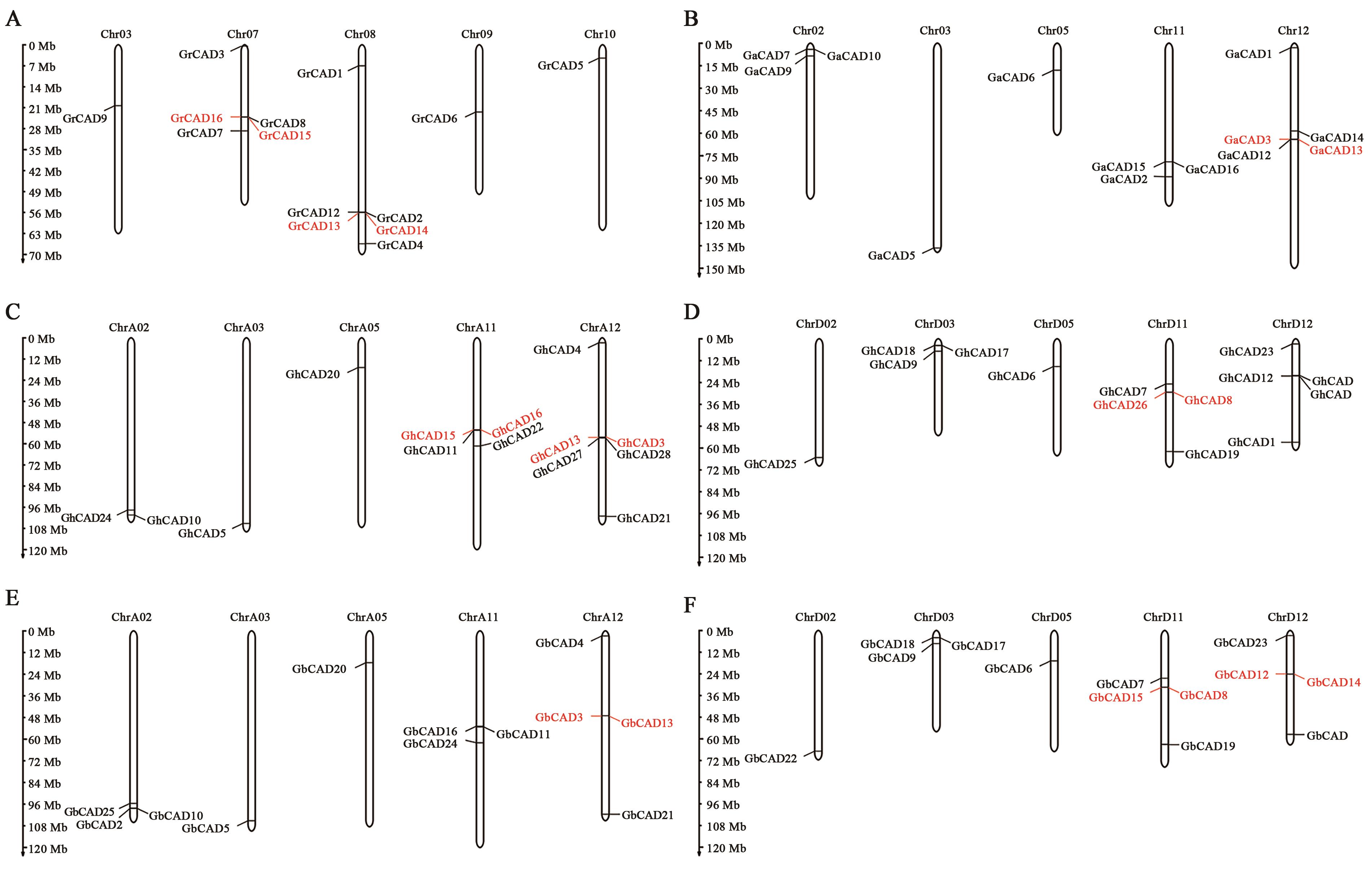
图1 雷蒙德氏棉、亚洲棉、陆地棉和海岛棉CAD基因家族的染色体定位A:雷蒙德氏棉CAD基因染色体定位;B:亚洲棉CAD基因染色体定位;C~D:陆地棉CAD基因染色体定位;E~F:海岛棉CAD基因染色体定位。红色表示串联重复序列。
Fig. 1 Genomic location of CAD genes on the G. raimondii, G. arboretum, G. hirsutum and G. barbadense chromosomes

图2 雷蒙德氏棉、亚洲棉、陆地棉和海岛棉CAD家族基因结构图A:雷蒙德氏棉CAD基因结构图;B:亚洲棉CAD基因结构图;C:陆地棉CAD基因结构图;D:海岛棉CAD基因结构图。外显子、非编码区和内含子分别用黑色方块、灰色方块和横线表示。
Fig. 2 Exons, introns and UTR analysis of G. raimondii, G. arboretum, G. hirsutum and G. barbadenseCAD gene family
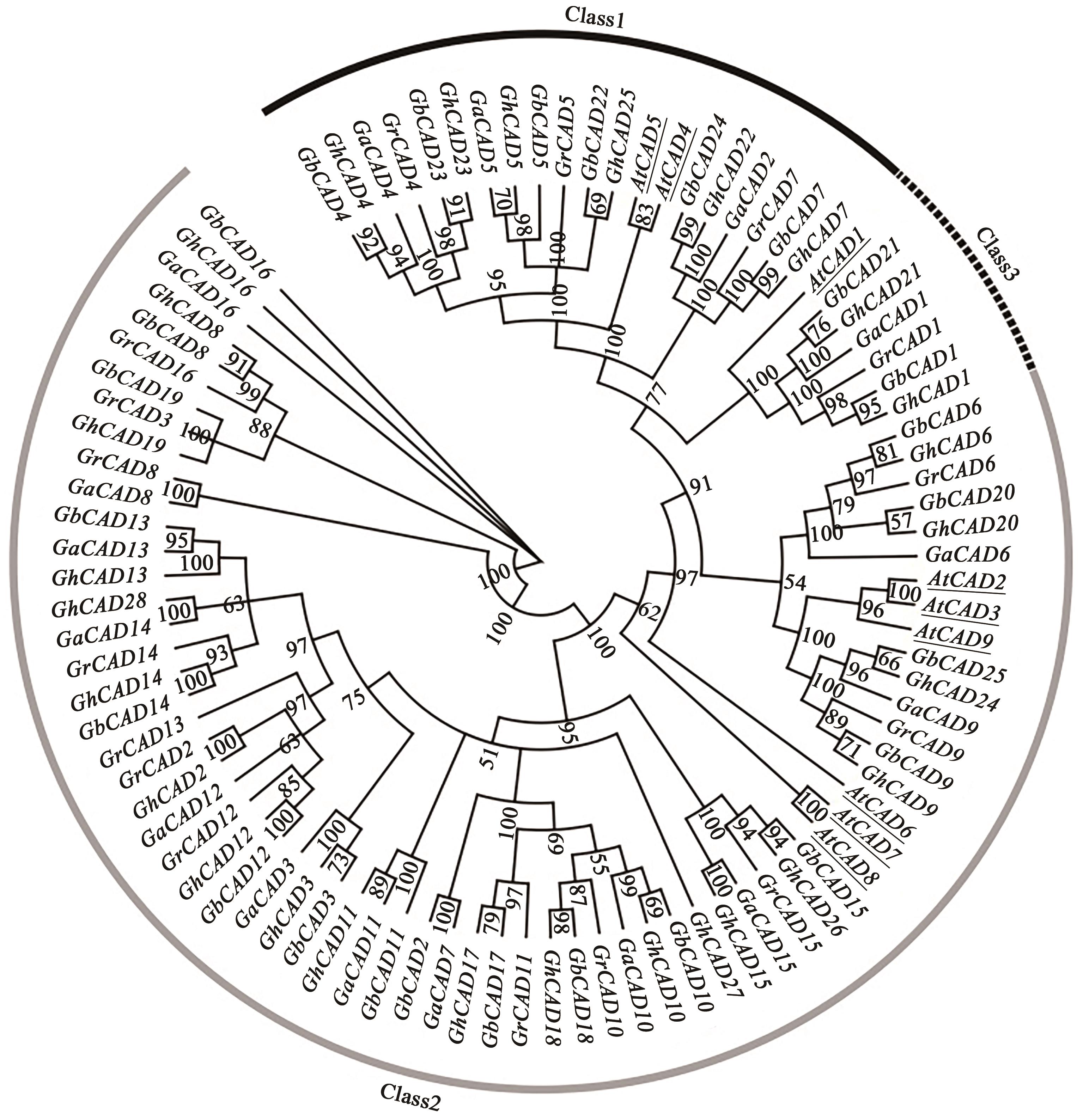
图3 棉花与拟南芥CAD基因家族的系统进化分析注:Class 1—棉花CAD基因组和拟南芥AtCAD4、AtCAD5聚为第1个亚类;Class 2—棉花CAD基因组和拟南芥AtCAD6、AtCAD7、AtCAD8聚为第2个亚类;Class 3—棉花CAD基因组和拟南芥AtCAD2、AtCAD3、AtCAD9聚为第3个亚类。带下划线表示拟南芥CAD基因。
Fig. 3 Phylogenetic analysis of CAD family genes in cotton and Arabidopsis
| 1 | BOERJAN W, RALPH J. BAUCHER M. Lignin biosynthesis[J]. Annu. Rev. Plant Biol., 2003, 54: 519-546. |
| 2 | GOU J Y, WANG L J, CHEN S P, et al.. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis[J]. Cell Res., 2007, 17: 422-434. |
| 3 | 尚军,吴旺泽,马永贵.植物苯丙烷代谢途径[J].中国生物化学与分子生物学报,2022,38(11):1467-1476. |
| 4 | FAN L, SHI W J, HU W R, et al.. Molecular and biochemical evidence for phenylpropanoid synthesis and presence of wall-linked phenolics in cotton fibers[J]. J. Integr. Plant Biol., 2009, 51(7): 626-637. |
| 5 | KIM S J, KIM M R, BEDGAR D L, et al.. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis [J]. Prco. Natl. Acad. Sci. USA, 2004, 101(6): 1455-1460. |
| 6 | TOBIAS C M, CHOW E K. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification[J]. Planta, 2005, 220(5): 678-688. |
| 7 | SABALLOS A, EJETA G, SANCHEZ E, et al.. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene[J]. Genetics, 2009, 181(2): 783-795. |
| 8 | KNIGHT M E, HALPIN C, SCHUCH W. Identification and characterization of cDNA clones encoding cinnamyl alcohol dehydrogenase from tobacco[J]. Plant Mol. Biol., 1992, 19(5): 793-801. |
| 9 | SIBOUT R, EUDES A, MOUILLE G, et al.. cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis [J]. Plant Cell, 2005, 17(7): 2059-2076. |
| 10 | KIM S J, KIM K W, CHO M H, et al.. Expression of cinnamyl alcohol dehydrogenase and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations?[J]. Phytochemistry, 2011, 68(14): 1957-1974. |
| 11 | LI L G, CHENG X F, LESHKEVICH J, et al.. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase[J]. Plant Cell, 2001, 13(7): 1567-1586. |
| 12 | GUILLAUMIE S, PICHON M, MARTINANT J P, et al. Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3 and bm4 young near-isogenic maize plants[J]. Planta, 2007, 226: 235-250. |
| 13 | ZHANG K, QIAN Q, HUANG Z, et al.. GOLD HULL AND INTERNODE2 encodes a primary multifunction cinnamyl-alcohol dehydrogenase in rice[J]. Plant Physiol., 2006, 140(3): 972-983. |
| 14 | GUO D M, RAN J H, WANG X Q. Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of bona fide CAD [J]. J. Mol. Evol., 2010, 71: 202-218. |
| 15 | 张鲁斌,谷会,弓德强,等.植物肉桂醇脱氢酶及其基因研究进展[J].西北植物学报,2011,31(1):204-211. |
| 16 | SCHUBERT R, SPERISEN C, MÜLLER-STARCK G, et al.. The cinnamyl alcohol dehydrogenase gene structure in Picea abies (L.)Karst: genomic sequences, southern hybridization, genetic analysis and phylogenetic relationships[J]. Trees, 1998, 12: 453-463. |
| 17 | QI K J, SONG X F, YUAN Y Z, et al.. CAD genes: genome-wide identification, evolution, and their contribution to lignin biosynthesis in pear (Pyrus bretschneideri)[J/OL]. Plants, 2021, 10: 1444[2021-07-15]. . |
| 18 | EL-GEBALI S, MISTRY J, BATEMAN A, et al.. The pfam protein families database in 2019[J]. Nucl. Acids Res., 2019, 47(D1): 427-432. |
| 19 | EOM H S, KIM H, HYUN K T. The cinnamyl alcohol dehydrogenase (CAD) gene family in flax (Linum usitatissimum L.): insight from expression profiling of cads induced by elicitors in cultured flax cells[J]. Arch. Biol. Sci., 2016, 68(3): 603-612. |
| 20 | 高正银,孙文杰,宋晓云,等.雷蒙德棉第Ⅲ类过氧化物酶全基因组鉴定和表达分析[J].生物技术进展,2019,9(5):490-501. |
| 21 | ZHANG T Z, HU Y, JIANG W K, et al.. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement[J]. Nat. Biotechnol., 2015, 33: 531-537. |
| 22 | CANNON S B, MITRA A, BAUMGARTEN A, et al.. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J/OL]. BMC Plant Biol., 2004, 4: 10[2004-06-01]. . |
| 23 | HISANO H, NANDAKUMAR R, WANG Z Y. Genetic modification of lignin biosynthesis for improved biofuel production[J]. In Vitro Cell Dev. Biol. Plant, 2009, 45: 306-313. |
| 24 | HOVAV R, UDALL J A, HOVAV E, et al.. A majority of cotton genes are expressed in single-celled fiber[J]. Planta, 2008, 227: 319-329. |
| 25 | HU W R, LINKER R, FAN L, et al.. Accumulation and structural unit of wall-linked phenolics in the cotton (Gossypium hirsutum) fiber secondary wall[J]. Int. J. Agric. Biol., 2019, 21: 1063-1072. |
| 26 | TRONCHET M, BALAGUÉ C, KROJ T, et al.. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis [J]. Mol. Plant Pathol., 2010, 11(1): 83-92. |
| 27 | SIBOUT R, EUDES A, POLLET B, et al.. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis isolation and characterization of the corresponding mutants[J]. Plant Physiol., 2003, 132: 848-860. |
| 28 | RAES J, ROHDE A, CHRISTENSEN J H, et al.. Genome-wide characterization of the lignification toolbox in Arabidopsis [J]. Plant Physiol., 2003, 133(3): 1051-1071. |
| 29 | LI X M, YUAN D J, ZHANG J F, et al.. Genetic mapping and characteristics of genes specifically or preferentially expressed during fiber development in cotton[J/OL]. PLoS ONE, 2013, 8: e54444[2013-01-25]. . |
| 30 | 胡文冉,李晓东,周小云,等.棉花GhCAD6基因表达及其功能分析研究[J].生物技术进展,2019, 9(1):46-53. |
| 31 | CAROCHA V, SOLER M, HEFER C, et al.. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis [J]. New Phytol., 2015, 206: 1297-1313. |
| 32 | HIRANO K, AYA K, KONDO M, et al.. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm[J]. Plant Cell Rep., 2012, 31(1): 91-101. |
| [1] | 朱兴墅, 黄军杰, 翁才明, 刘旺武, 陈永元, 陈惠燕, 赵虎, 王瑜, 林承志. 基于生物信息学分析林奇综合征相关结直肠癌关键Hub基因的筛选与功能富集分析[J]. 生物技术进展, 2023, 13(2): 291-297. |
| [2] | 李凤梅, 汤雪媛, 焦高洋, 齐玉亭, 杜献婷, 鲍倩倩, 张梦, 徐小博, 徐鑫, 张艳芳. 金银花花形基因的发掘及生物信息学分析[J]. 生物技术进展, 2023, 13(1): 90-101. |
| [3] | 于敏, 王敏, 魏延焕, 刘毅毅. SARS-CoV-2病毒感染潜在关键分子生物标志物及免疫浸润特征分析[J]. 生物技术进展, 2022, 12(5): 760-768. |
| [4] | 许广平,张志勇,任忠宏,张志伟. 黑鲷ELO基因编码蛋白结构与功能的生物信息学分析[J]. 生物技术进展, 2021, 11(3): 344-352. |
| [5] | 许杰,王可飞,魏晓晶,龚莉欣,焦阳,邱录贵,郝牧. 基于生物信息学分析SCHIP1在急性髓系白血病中的表达及其临床意义[J]. 生物技术进展, 2020, 10(4): 417-425. |
| [6] | 张凤丽,杨雅麟,夏锐,高辰辰,李栋,冉超,张震,张洪玲,周志刚. 吉陶单极虫可分泌蛋白酶基因的克隆与生物信息学分析[J]. 生物技术进展, 2019, 9(4): 375-383. |
| [7] | 徐长禄,赵艳红,马艺戈,王冰蕊,王鼎,郭青,佟静媛,高洁,李亚朴,刘金花,石莉红. 利用生物信息学方法解析自噬相关基因在人体红细胞终末分化各阶段的动态表达[J]. 生物技术进展, 2019, 9(3): 271-276. |
| [8] | 孙静娟,邱景璇,曾海娟,丁承超,王广彬,李杰,王淑娟,刘箐. 单增李斯特菌prsA1基因的截短表达、纯化及多克隆抗体的制备[J]. 生物技术进展, 2018, 8(3): 254-260. |
| [9] | 姜志磊,周琳,武奉慈,宋新元. 玉米扩展蛋白Expansin基因家族定位及基因表达模式分析[J]. 生物技术进展, 2018, 8(1): 34-40. |
| [10] | 张茂娜,姜亮,张焱. 硒代谢网络与硒蛋白质组的生物信息学研究进展[J]. 生物技术进展, 2017, 7(5): 537-544. |
| [11] | 康爽,贺红霞,王铭,郭嘉,薛晶. 杏蔗糖转运蛋白基因PaSUC4的获得及其生物信息学分析[J]. 生物技术进展, 2016, 6(2): 105-112. |
| [12] | 庞伟民,靳茜,王旭静,杨江涛,吕少溥,唐巧玲,王志兴. 陆地棉纤维优势表达基因GhRACK1的克隆与序列分析[J]. 生物技术进展, 2015, 5(5): 366-370. |
| [13] | 李圣彦,郎志宏,黄大昉. 真核生物启动子研究概述[J]. 生物技术进展, 2014, 4(3): 158-164. |
| [14] | 姚祖杰,吴松青,彭艳,杨梅. 一株高产杀虫蛋白苏云金杆菌的生物学特性研究以及其aiiA基因的克隆和生物信息学分析[J]. 生物技术进展, 2013, 3(2): 124-131. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部