


生物技术进展 ›› 2023, Vol. 13 ›› Issue (5): 730-741.DOI: 10.19586/j.2095-2341.2023.0106
蒲伟军1( ), 谭冰兰1, 贺丹晨1,2, 张盼1, 李玉斌1,3, 朱莉1(
), 谭冰兰1, 贺丹晨1,2, 张盼1, 李玉斌1,3, 朱莉1( )
)
收稿日期:2023-08-11
接受日期:2023-09-01
出版日期:2023-09-25
发布日期:2023-10-10
通讯作者:
朱莉
作者简介:蒲伟军 E-mail: pwj0613@163.com;
基金资助:
Weijun PU1( ), Binglan TAN1, Danchen HE1,2, Pan ZHANG1, Yubin LI1,3, Li ZHU1(
), Binglan TAN1, Danchen HE1,2, Pan ZHANG1, Yubin LI1,3, Li ZHU1( )
)
Received:2023-08-11
Accepted:2023-09-01
Online:2023-09-25
Published:2023-10-10
Contact:
Li ZHU
摘要:
高粱是重要的禾谷类作物,其测序品种BTx623的全基因组序列已经公布,为高粱InDel 标记的开发提供了研究基础。插入与缺失(insertion/deletion,InDel)标记作为高通量分子标记,在植物基因组中具有遗传稳定性高、分布广、多态性强、通用性强等优点,目前已在水稻、玉米、棉花等主要作物中得到广泛应用,然而在高粱中的研究尚不多见。以高粱品种JIUTIAN1基因组DNA为研究对象,利用Illumina平台进行重测序,进一步利用Trimmomatic软件对测序原始数据进行质量控制,剔除低质量数据,使用BWA软件将获得的高质量有效数据与测序品种BTx623参考基因组进行序列比对,从中检测出大量InDel位点。利用这些InDel位点在全基因组范围内设计均匀分布的205个标记,通过PCR验证后筛选出87个多态性标记,多态性为42.44%。高粱Indel标记的开发有望为高粱遗传连锁图谱的构建、遗传多样性分析、杂交种纯度鉴定、高粱重要农艺性状相关基因的定位和分子标记辅助选择育种等研究奠定基础。
中图分类号:
蒲伟军, 谭冰兰, 贺丹晨, 张盼, 李玉斌, 朱莉. 利用重测序技术开发高粱InDel分子标记[J]. 生物技术进展, 2023, 13(5): 730-741.
Weijun PU, Binglan TAN, Danchen HE, Pan ZHANG, Yubin LI, Li ZHU. Development of InDel Molecular Markers in Sorghum Using Re-sequencing Technology[J]. Current Biotechnology, 2023, 13(5): 730-741.
| 编号 | 引物名称 | 引物序列 (5'→3') | 编号 | 引物名称 | 引物序列 (5'→3') |
|---|---|---|---|---|---|
| InDel01 | Chr01-NP2-F | 5'-TGGTCGAACAAGACTGCAAC-3' | InDel45 | Chr05-NP8-F | 5'-TTGTGGGATGAAAGGAGCAT-3' |
| Chr01-NP2-R | 5'-CACTACGGCGAAATTGGAAG-3' | Chr05-NP8-R | 5'-GCCTGTACCAGAAACCGAAC-3' | ||
| InDel02 | Chr01-NP3-F | 5'-ATCGGCATCCATCAGATTTT-3' | InDel46 | Chr05-NP9-F | 5'-AGAATGTTGCACGCATTGAC-3' |
| Chr01-NP3-R | 5'-CCTGCCAACTCAGCAATTTT-3' | Chr05-NP9-R | 5'-GCCGACCATTATTTGTGACC-3' | ||
| InDel03 | Chr01-NP4-F | 5'-TCTTTCCCAGTGGAACAACC-3' | InDel47 | Chr05-NP10-F | 5'-GCAATTGCAACATCGAACAC-3' |
| Chr01-NP4-R | 5'-CAGGGGACTCTTCAAACTGC-3' | Chr05-NP10-R | 5'-TCCACATCCATCCTCATCAA-3' | ||
| InDel04 | Chr01-NP6-F | 5'-AGATCCGAGCATTTCTGCTG-3' | InDel48 | Chr06-MP1-F | 5'-CATAGGATTTTGCCACGACA-3' |
| Chr01-NP6-R | 5'-AGTTCGCCCAGGTAAGGTCT-3' | Chr06-MP1-R | 5'-CGTCACAGAGTCCATGCAGA-3' | ||
| InDel05 | Chr02-NP2-F | 5'-TTTCATGGCTTTGAGTGTCA-3' | InDel49 | Chr06-MP8-F | 5'-TGCGAAAAGAAAAAGGATGG-3' |
| Chr02-NP2-R | 5'-ACGCTAACAGAGGGCAGTGT-3' | Chr06-MP8-R | 5'-CCCAAAATCCACTTTCCTGA-3' | ||
| InDel06 | Chr02-NP4-F | 5'-TGGTCAAGTTTTGGCATGAA-3' | InDel50 | Chr06-MP12-F | 5'-TACGCTGGTGCTTACGAAAA-3' |
| Chr02-NP4-R | 5'-GTCATGTTCTTTCTGATTCCCTCT-3' | Chr06-MP12-R | 5'-GGGAGAGGAAAGGTGAAACC-3' | ||
| InDel07 | Chr02-NP7-F | 5'-TGCCTCTATTTTTCCCATGC-3' | InDel51 | Chr06-NP2-F | 5'-ATTGATTTGGGGTCGGGTA-3' |
| Chr02-NP7-R | 5'-AATGCCAAGTTTGGTCCATC-3' | Chr06-NP2-R | 5'-CACCGCAAGCCAAAATAGAG-3' | ||
| InDel08 | Chr02-NP10-F | 5'-TACACACAGTACGCCCATGC-3' | InDel52 | Chr06-NP7-F | 5'-AGTGGCTGTACGGGTTTGAC-3' |
| Chr02-NP10-R | 5'-GGTTTGGTGTCTGGCAACTT-3' | Chr06-NP7-R | 5'-TTGTGCAGCATCAACGTGTA-3' | ||
| InDel09 | Chr02-NP11-F | 5'-ATATCTGGTTCCCCCAGCTT-3' | InDel53 | Chr07-NP1-F | 5'-TGGATCCAAGTAAACCCGATA-3' |
| Chr02-NP11-R | 5'-GCTCAGCTCGTCTCGTCTCT-3' | Chr07-NP1-R | 5'-ACTTAAAGGCCCCACTGAAA-3' | ||
| InDel10 | Chr02-NP12-F | 5'-TTTTGCGACACCTACTGCTG-3' | InDel54 | Chr07-NP3-F | 5'-TAACGGTTTGGGTCTTCGAG-3' |
| Chr02-NP12-R | 5'-GGTTTTCTGGGGTGCATAGA-3' | Chr07-NP3-R | 5'-GCGATGAAAAACACAGCAAA-3' | ||
| InDel11 | Chr02-NP14-F | 5'-TGCAATACCTGTTCCCCACT-3' | InDel55 | Chr07-NP6-F | 5'-TTCTGGTCTGGTGAATGCAA-3' |
| Chr02-NP14-R | 5'-CAACAATGGCTACAGCAACG-3' | Chr07-NP6-R | 5'-TCCATGACGAACGAGAAATC-3' | ||
| InDel12 | Chr02-MP1-F | 5'-CCTGACCCAGACACACACAC-3' | InDel56 | Chr07-MP8-F | 5'-GCCAAGTTTGCCAAAATGC-3' |
| Chr02-MP1-R | 5'-AGCAATCTTGTCAGGGGATG-3' | Chr07-MP8-R | 5'-GTTAACGCAACCCAGCAGA-3' | ||
| InDel13 | Chr02-MP2-F | 5'-GCTGATGGCTCTCCAAACTC-3' | InDel57 | Chr08-MP1-F | 5'-GCCGAGTGTCTCGTTAGACC-3' |
| Chr02-MP2-R | 5'-GTTGGTCCCTGAATGCTTGT-3' | Chr08-MP1-R | 5'-CACGACGGCACCCTACTAAT-3' | ||
| InDel14 | Chr02-MP4-F | 5'-TGACCTAAGGGGTTGTTTGG-3' | InDel58 | Chr08-MP12-F | 5'-GAACCAAACAGCGTCAGACC-3' |
| Chr02-MP4-R | 5'-GCACCTGCCACATCCTATTT-3' | Chr08-MP12-R | 5'-AGCACCACCCATAACGTGTC-3' | ||
| InDel15 | Chr02-MP12-F | 5'-CAAAGGCCGATGTATTTGCT-3' | InDel59 | Chr08-AP2-F | 5'-TGACTGTGTCGCTACCTTGC-3' |
| Chr02-MP12-R | 5'-TGTGCCAACCCTGTATGTGT-3' | Chr08-AP2-R | 5'-GATGGCAAGAGTGGGATTTG-3' | ||
| InDel16 | Chr02-MP13-F | 5'-GAAGGCGCTCGTGAATTTTA-3' | InDel60 | Chr08-AP4-F | 5'-TCCAAACCGGTGGATAATTG-3' |
| Chr02-MP13-R | 5'-CAGACACAAAGGCTGTCTCG-3' | Chr08-AP4-R | 5'-AAGCCTTGTTTGGATGCTGT-3' | ||
| InDel17 | Chr02-MP14-F | 5'-CCGCACAAAGTAGAGCATCA-3' | InDel61 | Chr08-AP5-F | 5'-CAAAGGACGAGATAAGCA-3' |
| Chr02-MP14-R | 5'-CCGTAAGAGAAGCGAGCCTA-3' | Chr08-AP5-R | 5'-TTCCCTGTCACATCAAAT-3' | ||
| InDel18 | Chr02-MP16-F | 5'-TTTTCAGTCTGGTCGCTTGA-3' | InDel62 | Chr08-NP3-F | 5'-TCCATGCATATGCCTCAAAA-3' |
| Chr02-MP16-R | 5'-CACACCTCACTCGCTTCTCA-3' | Chr08-NP3-R | 5'-CAAGTTGTGCAGTTGCTGGT-3' | ||
| InDel19 | Chr03-MP3-F | 5'-GGCACATGCATGAAGCACTA-3' | InDel63 | Chr09-MP1-F | 5'-GACGGAGATACTTAGGGCAGAA-3' |
| Chr03-MP3-R | 5'-TGATTCGACAGCTTCCAGAG-3' | Chr09-MP1-R | 5'-TCTCTCTCCTCCCTTTCCAA-3' | ||
| InDel20 | Chr03-MP5-F | 5'-CGAGAAGACATGACAAAACGAG-3' | InDel64 | Chr09-YM1-F | 5'-GTCGGCGTAGCTCATCTT-3' |
| Chr03-MP5-R | 5'-CTTTGGTCGCATGTATGGAA-3' | Chr09-YM1-R | 5'-GTAGCACTTTCGTTTGTATTTG-3' | ||
| InDel21 | Chr03-MP10-F | 5'-CCATCTGGATATACACGGGTTAG-3' | InDel65 | Chr09-YM2-F | 5'-GTCAAGGAGGAACCAAAG-3' |
| Chr03-MP10-R | 5'-TGCAAACCGGTTCTCAAAGT-3' | Chr09-YM2-R | 5'-ATGCCTAAGCCTAAGTAA-3' | ||
| InDel22 | Chr03-MP12-F | 5'-TGCATGCACGTAGTAGACACA-3' | InDel66 | Chr09-YM5-F | 5'-GTCAAGGAGGAACCAAAG-3' |
| Chr03-MP12-R | 5'-CACCACGATAGCAAATCACC-3' | Chr09-YM5-R | |||
| InDel23 | Chr03-MP14-F | 5'-GGTGCAGTGGTGTCTTCCTT-3' | InDel67 | Chr09-YM10-F | 5'-TCGGCTTGATTTGTTCTT-3' |
| Chr03-MP14-R | 5'-ATTTACAGCGCGTGGCTATC-3' | Chr09-YM10-R | 5'-CTGGCTGCTGAGTAAGTG-3' | ||
| InDel24 | Chr03-NP3-F | 5'-TTAAGGTCTCGTTTAGTTCAGG-3' | InDel68 | Chr09-YM11-F | 5'-CGAAATGCTTCACGCTGT-3' |
| Chr03-NP3-R | 5'-GCCTTGTTCAGTTGGCAAAA-3' | Chr09-YM11-R | 5'-GAAACCTACCCAATGTCCG-3' | ||
| InDel25 | Chr03-NP4-F | 5'-TCAGATGAGCAGGCTCTACAGT-3' | InDel69 | Chr09-MP4-F | 5'-TTCCAATAAAGCTGCCGTTC-3' |
| Chr03-NP4-R | 5'-GAGCATTTTCGGCATGTGTA-3' | Chr09-MP4-R | 5'-ATACTAGCGGTGCGGGTTAC-3' | ||
| InDel26 | Chr03-NP7-F | 5'-ACGAAAGAAATGCACCGAAC-3' | InDel70 | Chr09-MP5-F | 5'-CTGAACAAGGCCTAGGTGGA-3' |
| Chr03-NP7-R | 5'-CCCAGCCTAGGAAACATCAA-3' | Chr09-MP5-R | 5'-GCTCACCACTGCAACTGCTA-3' | ||
| InDel27 | Chr03-NP9-F | 5'-GCGTGACTATTGCAGCTGTT-3' | InDel71 | Chr09-MP9-F | 5'-ATGTCAGCGAAATGGTTTCC-3' |
| Chr03-NP9-R | 5'-CATGCCATCCACTTCCACTA-3' | Chr09-MP9-R | 5'-CACCAAGTCCTTGCTCATCA-3' | ||
| InDel28 | Chr04-MP1-F | 5'-GCGGTCATAGGCGTTACTGT-3' | InDel72 | Chr09-MP11-F | 5'-ATGGAATATGAGCCCCAACA-3' |
| Chr04-MP1-R | 5'-CGCTGTCACATCGSSTCTTT-3' | Chr09-MP11-R | 5'-ATCAGGTCGTCCACCAGTTC-3' | ||
| InDel29 | Chr04-MP2-F | 5'-TGGATAGGTTCATGCTGCTG-3' | InDel73 | Chr09-NP1-F | 5'-ACGAGGATGGCAAGAGGATA-3' |
| Chr04-MP2-R | 5'-TTTTTGCCACCTTCCAATTC-3' | Chr09-NP1-R | 5'-GACGGTGCATGTGCTTTTT-3' | ||
| InDel30 | Chr04-MP3-F | 5'-CTGCCCTATGCATCTGTTCA-3' | InDel74 | Chr09-NP2-F | 5'-GCGGATGAGGAGGAATCTCT-3' |
| Chr04-MP3-R | 5'-CCAACTAGTTCAGGGCAGGA-3' | Chr09-NP2-R | 5'-TTTACTGCGCCTCGTCTCTT-3' | ||
| InDel31 | Chr04-MP13-F | 5'-TCCGTCCAACTGGAATTGAT-3' | InDel75 | Chr09-NP4-F | 5'-AGCACTAGCAAATGCCACAA-3' |
| Chr04-MP13-R | 5'-AATCCTTCGTGGAAAGCTGA-3' | Chr09-NP4-R | 5'-ATGACGTTTTTCGGCTTGTC-3' | ||
| InDel32 | Chr04-NP1-F | 5'-ATCTTGCGGCACATACATGA-3' | InDel76 | Chr09-NP5-F | 5'-GCTGAGCACTTGAGGGTTGT-3' |
| Chr04-NP1-R | 5'-CTTCAACACTCAGCCCACCT-3' | Chr09-NP5-R | 5'-CTGGTCATCGGCTTAGCAAT-3' | ||
| InDel33 | Chr04-NP2-F | 5'-TATAAAGCACCGGTGGAGGT-3' | InDel77 | Chr09-NP9-F | 5'-GCGGCTTATCACAGCTCCTA-3' |
| Chr04-NP2-R | 5'-ACAAATCGTTTCACCGCTTC-3' | Chr09-NP9-R | 5'-TCACCATGCAAAACGTCCTA-3' | ||
| InDel34 | Chr04-NP6-F | 5'-GGCTGTTGGAGGAATTTTGA-3' | InDel78 | Chr10-MP2-F | 5'-AGAGATCGAGAGGACGGACA-3' |
| Chr04-NP6-R | 5'-CCTTGTACTCTTCTGAGTTTTGGA-3' | Chr10-MP2-R | 5'-GGAATTCCATGCCCAAATC-3' | ||
| InDel35 | Chr04-NP7-F | 5'-GACCGAGAAAGAAAATCATACACTC-3' | InDel79 | Chr10-MP5-F | 5'-GCATTTGCATCTGAAAGCTC-3' |
| Chr04-NP7-R | 5'-AGGCAAAAAGCCCAACTCTT-3' | Chr10-MP5-R | 5'-ACCAAACTCTGCAGGAATGC-3' | ||
| InDel36 | Chr04-NP8-F | 5'-ATGCCGTAGATCGTCCTCCT-3' | InDel80 | Chr10-MP7-F | 5'-AGGCGTTGCCTCTAACACTG-3' |
| Chr04-NP8-R | 5'-CAACCGGAGCTGGAAATG-3' | Chr10-MP7-R | 5'-GTCCGGTGAGTTCGTTTCTC-3' | ||
| InDel37 | Chr04-NP11-F | 5'-AGCTCATCAAAGCGAAAGGA-3' | InDel81 | Chr10-MP8-F | 5'-CACATGGTTCTTTTCCACGA-3' |
| Chr04-NP11-R | 5'-AGTCTAGACGCCTCGACGTT-3' | Chr10-MP8-R | 5'-GGAGTTGGATGAAAGCCTTG-3' | ||
| InDel38 | Chr05-MP9-F | 5'-ATGGGAAGGCCTACGATTTT-3' | InDel82 | Chr10-MP11-F | 5'-TGGTAGACGGGTCCTAGTGC-3' |
| Chr05-MP9-R | 5'-TGGGTGATGGTAGGAGAATAGA-3' | Chr10-MP11-R | 5'-TACATCGCTGCTGGAGTGTC-3' | ||
| InDel39 | Chr05-MP13-F | 5'-ATTTAACCTGGCTGCCTGTC-3' | InDel83 | Chr10-NP4-F | 5'-ACACGGGTCGAAGATCAAAG-3' |
| Chr05-MP13-R | 5'-GACCGCGACACTAACAAATG-3' | Chr10-NP4-R | 5'-GGTGCCAACTTCCTTCATCA-3' | ||
| InDel40 | Chr05-MP14-F | 5'-TTTGCCTAGCCATTTCAGGT-3' | InDel84 | Chr10-NP6-F | 5'-TTTGCAAGCTGTTTTGGTTC-3' |
| Chr05-MP14-R | 5'-CACGGTCAACAATCAGCTCT-3' | Chr10-NP6-R | 5'-GGGTGAAGGCTACATCGGTA-3' | ||
| InDel41 | Chr05-NP1-F | 5'-AGCATTGGCTGCTAAGCATT-3' | InDel85 | Chr10-NP7-F | 5'-TACCGTTGTCCGACGCTAAT-3' |
| Chr05-NP1-R | 5'-TAGCAGCAACAGCAAACAGG-3' | Chr10-NP7-R | 5'-TTGTGTAGGCGTCGTGCTTA-3' | ||
| InDel42 | Chr05-NP2-F | 5'-AGAGAGAATCCTCGGCAAGA-3' | InDel86 | Chr10-NP9-F | 5'-CGACCCTGTCGTATGTATGC-3' |
| Chr05-NP2-R | 5'-TTTGTGAGGGGTTCTTGAGC-3' | Chr10-NP9-R | 5'-TTGTCCTTCTGTGGCTGTTG-3' | ||
| InDel43 | Chr05-NP5-F | 5'-CTTCGTGGACCACATGACAC-3' | InDel87 | Chr10-NP11-F | 5'-ACGGAATGCAAAAAGCAAAG-3' |
| Chr05-NP5-R | 5'-GTACCCTCGCCAGTCAACTC-3' | Chr10-NP11-R | 5'-GGTGACTTCAACCGGGATTA-3' | ||
| InDel44 | Chr05-NP6-F | 5'-TCTCACGGGTCGAAAGTACC-3' | |||
| Chr05-NP6-R | 5'-CAATCTCGATTGCCTCAACA-3' |
表 1 分布于 10 条染色体上的 87 对 InDel 引物信息
Table 1 Information of 87 pairs of InDel primers distributed on 10 chromosomes
| 编号 | 引物名称 | 引物序列 (5'→3') | 编号 | 引物名称 | 引物序列 (5'→3') |
|---|---|---|---|---|---|
| InDel01 | Chr01-NP2-F | 5'-TGGTCGAACAAGACTGCAAC-3' | InDel45 | Chr05-NP8-F | 5'-TTGTGGGATGAAAGGAGCAT-3' |
| Chr01-NP2-R | 5'-CACTACGGCGAAATTGGAAG-3' | Chr05-NP8-R | 5'-GCCTGTACCAGAAACCGAAC-3' | ||
| InDel02 | Chr01-NP3-F | 5'-ATCGGCATCCATCAGATTTT-3' | InDel46 | Chr05-NP9-F | 5'-AGAATGTTGCACGCATTGAC-3' |
| Chr01-NP3-R | 5'-CCTGCCAACTCAGCAATTTT-3' | Chr05-NP9-R | 5'-GCCGACCATTATTTGTGACC-3' | ||
| InDel03 | Chr01-NP4-F | 5'-TCTTTCCCAGTGGAACAACC-3' | InDel47 | Chr05-NP10-F | 5'-GCAATTGCAACATCGAACAC-3' |
| Chr01-NP4-R | 5'-CAGGGGACTCTTCAAACTGC-3' | Chr05-NP10-R | 5'-TCCACATCCATCCTCATCAA-3' | ||
| InDel04 | Chr01-NP6-F | 5'-AGATCCGAGCATTTCTGCTG-3' | InDel48 | Chr06-MP1-F | 5'-CATAGGATTTTGCCACGACA-3' |
| Chr01-NP6-R | 5'-AGTTCGCCCAGGTAAGGTCT-3' | Chr06-MP1-R | 5'-CGTCACAGAGTCCATGCAGA-3' | ||
| InDel05 | Chr02-NP2-F | 5'-TTTCATGGCTTTGAGTGTCA-3' | InDel49 | Chr06-MP8-F | 5'-TGCGAAAAGAAAAAGGATGG-3' |
| Chr02-NP2-R | 5'-ACGCTAACAGAGGGCAGTGT-3' | Chr06-MP8-R | 5'-CCCAAAATCCACTTTCCTGA-3' | ||
| InDel06 | Chr02-NP4-F | 5'-TGGTCAAGTTTTGGCATGAA-3' | InDel50 | Chr06-MP12-F | 5'-TACGCTGGTGCTTACGAAAA-3' |
| Chr02-NP4-R | 5'-GTCATGTTCTTTCTGATTCCCTCT-3' | Chr06-MP12-R | 5'-GGGAGAGGAAAGGTGAAACC-3' | ||
| InDel07 | Chr02-NP7-F | 5'-TGCCTCTATTTTTCCCATGC-3' | InDel51 | Chr06-NP2-F | 5'-ATTGATTTGGGGTCGGGTA-3' |
| Chr02-NP7-R | 5'-AATGCCAAGTTTGGTCCATC-3' | Chr06-NP2-R | 5'-CACCGCAAGCCAAAATAGAG-3' | ||
| InDel08 | Chr02-NP10-F | 5'-TACACACAGTACGCCCATGC-3' | InDel52 | Chr06-NP7-F | 5'-AGTGGCTGTACGGGTTTGAC-3' |
| Chr02-NP10-R | 5'-GGTTTGGTGTCTGGCAACTT-3' | Chr06-NP7-R | 5'-TTGTGCAGCATCAACGTGTA-3' | ||
| InDel09 | Chr02-NP11-F | 5'-ATATCTGGTTCCCCCAGCTT-3' | InDel53 | Chr07-NP1-F | 5'-TGGATCCAAGTAAACCCGATA-3' |
| Chr02-NP11-R | 5'-GCTCAGCTCGTCTCGTCTCT-3' | Chr07-NP1-R | 5'-ACTTAAAGGCCCCACTGAAA-3' | ||
| InDel10 | Chr02-NP12-F | 5'-TTTTGCGACACCTACTGCTG-3' | InDel54 | Chr07-NP3-F | 5'-TAACGGTTTGGGTCTTCGAG-3' |
| Chr02-NP12-R | 5'-GGTTTTCTGGGGTGCATAGA-3' | Chr07-NP3-R | 5'-GCGATGAAAAACACAGCAAA-3' | ||
| InDel11 | Chr02-NP14-F | 5'-TGCAATACCTGTTCCCCACT-3' | InDel55 | Chr07-NP6-F | 5'-TTCTGGTCTGGTGAATGCAA-3' |
| Chr02-NP14-R | 5'-CAACAATGGCTACAGCAACG-3' | Chr07-NP6-R | 5'-TCCATGACGAACGAGAAATC-3' | ||
| InDel12 | Chr02-MP1-F | 5'-CCTGACCCAGACACACACAC-3' | InDel56 | Chr07-MP8-F | 5'-GCCAAGTTTGCCAAAATGC-3' |
| Chr02-MP1-R | 5'-AGCAATCTTGTCAGGGGATG-3' | Chr07-MP8-R | 5'-GTTAACGCAACCCAGCAGA-3' | ||
| InDel13 | Chr02-MP2-F | 5'-GCTGATGGCTCTCCAAACTC-3' | InDel57 | Chr08-MP1-F | 5'-GCCGAGTGTCTCGTTAGACC-3' |
| Chr02-MP2-R | 5'-GTTGGTCCCTGAATGCTTGT-3' | Chr08-MP1-R | 5'-CACGACGGCACCCTACTAAT-3' | ||
| InDel14 | Chr02-MP4-F | 5'-TGACCTAAGGGGTTGTTTGG-3' | InDel58 | Chr08-MP12-F | 5'-GAACCAAACAGCGTCAGACC-3' |
| Chr02-MP4-R | 5'-GCACCTGCCACATCCTATTT-3' | Chr08-MP12-R | 5'-AGCACCACCCATAACGTGTC-3' | ||
| InDel15 | Chr02-MP12-F | 5'-CAAAGGCCGATGTATTTGCT-3' | InDel59 | Chr08-AP2-F | 5'-TGACTGTGTCGCTACCTTGC-3' |
| Chr02-MP12-R | 5'-TGTGCCAACCCTGTATGTGT-3' | Chr08-AP2-R | 5'-GATGGCAAGAGTGGGATTTG-3' | ||
| InDel16 | Chr02-MP13-F | 5'-GAAGGCGCTCGTGAATTTTA-3' | InDel60 | Chr08-AP4-F | 5'-TCCAAACCGGTGGATAATTG-3' |
| Chr02-MP13-R | 5'-CAGACACAAAGGCTGTCTCG-3' | Chr08-AP4-R | 5'-AAGCCTTGTTTGGATGCTGT-3' | ||
| InDel17 | Chr02-MP14-F | 5'-CCGCACAAAGTAGAGCATCA-3' | InDel61 | Chr08-AP5-F | 5'-CAAAGGACGAGATAAGCA-3' |
| Chr02-MP14-R | 5'-CCGTAAGAGAAGCGAGCCTA-3' | Chr08-AP5-R | 5'-TTCCCTGTCACATCAAAT-3' | ||
| InDel18 | Chr02-MP16-F | 5'-TTTTCAGTCTGGTCGCTTGA-3' | InDel62 | Chr08-NP3-F | 5'-TCCATGCATATGCCTCAAAA-3' |
| Chr02-MP16-R | 5'-CACACCTCACTCGCTTCTCA-3' | Chr08-NP3-R | 5'-CAAGTTGTGCAGTTGCTGGT-3' | ||
| InDel19 | Chr03-MP3-F | 5'-GGCACATGCATGAAGCACTA-3' | InDel63 | Chr09-MP1-F | 5'-GACGGAGATACTTAGGGCAGAA-3' |
| Chr03-MP3-R | 5'-TGATTCGACAGCTTCCAGAG-3' | Chr09-MP1-R | 5'-TCTCTCTCCTCCCTTTCCAA-3' | ||
| InDel20 | Chr03-MP5-F | 5'-CGAGAAGACATGACAAAACGAG-3' | InDel64 | Chr09-YM1-F | 5'-GTCGGCGTAGCTCATCTT-3' |
| Chr03-MP5-R | 5'-CTTTGGTCGCATGTATGGAA-3' | Chr09-YM1-R | 5'-GTAGCACTTTCGTTTGTATTTG-3' | ||
| InDel21 | Chr03-MP10-F | 5'-CCATCTGGATATACACGGGTTAG-3' | InDel65 | Chr09-YM2-F | 5'-GTCAAGGAGGAACCAAAG-3' |
| Chr03-MP10-R | 5'-TGCAAACCGGTTCTCAAAGT-3' | Chr09-YM2-R | 5'-ATGCCTAAGCCTAAGTAA-3' | ||
| InDel22 | Chr03-MP12-F | 5'-TGCATGCACGTAGTAGACACA-3' | InDel66 | Chr09-YM5-F | 5'-GTCAAGGAGGAACCAAAG-3' |
| Chr03-MP12-R | 5'-CACCACGATAGCAAATCACC-3' | Chr09-YM5-R | |||
| InDel23 | Chr03-MP14-F | 5'-GGTGCAGTGGTGTCTTCCTT-3' | InDel67 | Chr09-YM10-F | 5'-TCGGCTTGATTTGTTCTT-3' |
| Chr03-MP14-R | 5'-ATTTACAGCGCGTGGCTATC-3' | Chr09-YM10-R | 5'-CTGGCTGCTGAGTAAGTG-3' | ||
| InDel24 | Chr03-NP3-F | 5'-TTAAGGTCTCGTTTAGTTCAGG-3' | InDel68 | Chr09-YM11-F | 5'-CGAAATGCTTCACGCTGT-3' |
| Chr03-NP3-R | 5'-GCCTTGTTCAGTTGGCAAAA-3' | Chr09-YM11-R | 5'-GAAACCTACCCAATGTCCG-3' | ||
| InDel25 | Chr03-NP4-F | 5'-TCAGATGAGCAGGCTCTACAGT-3' | InDel69 | Chr09-MP4-F | 5'-TTCCAATAAAGCTGCCGTTC-3' |
| Chr03-NP4-R | 5'-GAGCATTTTCGGCATGTGTA-3' | Chr09-MP4-R | 5'-ATACTAGCGGTGCGGGTTAC-3' | ||
| InDel26 | Chr03-NP7-F | 5'-ACGAAAGAAATGCACCGAAC-3' | InDel70 | Chr09-MP5-F | 5'-CTGAACAAGGCCTAGGTGGA-3' |
| Chr03-NP7-R | 5'-CCCAGCCTAGGAAACATCAA-3' | Chr09-MP5-R | 5'-GCTCACCACTGCAACTGCTA-3' | ||
| InDel27 | Chr03-NP9-F | 5'-GCGTGACTATTGCAGCTGTT-3' | InDel71 | Chr09-MP9-F | 5'-ATGTCAGCGAAATGGTTTCC-3' |
| Chr03-NP9-R | 5'-CATGCCATCCACTTCCACTA-3' | Chr09-MP9-R | 5'-CACCAAGTCCTTGCTCATCA-3' | ||
| InDel28 | Chr04-MP1-F | 5'-GCGGTCATAGGCGTTACTGT-3' | InDel72 | Chr09-MP11-F | 5'-ATGGAATATGAGCCCCAACA-3' |
| Chr04-MP1-R | 5'-CGCTGTCACATCGSSTCTTT-3' | Chr09-MP11-R | 5'-ATCAGGTCGTCCACCAGTTC-3' | ||
| InDel29 | Chr04-MP2-F | 5'-TGGATAGGTTCATGCTGCTG-3' | InDel73 | Chr09-NP1-F | 5'-ACGAGGATGGCAAGAGGATA-3' |
| Chr04-MP2-R | 5'-TTTTTGCCACCTTCCAATTC-3' | Chr09-NP1-R | 5'-GACGGTGCATGTGCTTTTT-3' | ||
| InDel30 | Chr04-MP3-F | 5'-CTGCCCTATGCATCTGTTCA-3' | InDel74 | Chr09-NP2-F | 5'-GCGGATGAGGAGGAATCTCT-3' |
| Chr04-MP3-R | 5'-CCAACTAGTTCAGGGCAGGA-3' | Chr09-NP2-R | 5'-TTTACTGCGCCTCGTCTCTT-3' | ||
| InDel31 | Chr04-MP13-F | 5'-TCCGTCCAACTGGAATTGAT-3' | InDel75 | Chr09-NP4-F | 5'-AGCACTAGCAAATGCCACAA-3' |
| Chr04-MP13-R | 5'-AATCCTTCGTGGAAAGCTGA-3' | Chr09-NP4-R | 5'-ATGACGTTTTTCGGCTTGTC-3' | ||
| InDel32 | Chr04-NP1-F | 5'-ATCTTGCGGCACATACATGA-3' | InDel76 | Chr09-NP5-F | 5'-GCTGAGCACTTGAGGGTTGT-3' |
| Chr04-NP1-R | 5'-CTTCAACACTCAGCCCACCT-3' | Chr09-NP5-R | 5'-CTGGTCATCGGCTTAGCAAT-3' | ||
| InDel33 | Chr04-NP2-F | 5'-TATAAAGCACCGGTGGAGGT-3' | InDel77 | Chr09-NP9-F | 5'-GCGGCTTATCACAGCTCCTA-3' |
| Chr04-NP2-R | 5'-ACAAATCGTTTCACCGCTTC-3' | Chr09-NP9-R | 5'-TCACCATGCAAAACGTCCTA-3' | ||
| InDel34 | Chr04-NP6-F | 5'-GGCTGTTGGAGGAATTTTGA-3' | InDel78 | Chr10-MP2-F | 5'-AGAGATCGAGAGGACGGACA-3' |
| Chr04-NP6-R | 5'-CCTTGTACTCTTCTGAGTTTTGGA-3' | Chr10-MP2-R | 5'-GGAATTCCATGCCCAAATC-3' | ||
| InDel35 | Chr04-NP7-F | 5'-GACCGAGAAAGAAAATCATACACTC-3' | InDel79 | Chr10-MP5-F | 5'-GCATTTGCATCTGAAAGCTC-3' |
| Chr04-NP7-R | 5'-AGGCAAAAAGCCCAACTCTT-3' | Chr10-MP5-R | 5'-ACCAAACTCTGCAGGAATGC-3' | ||
| InDel36 | Chr04-NP8-F | 5'-ATGCCGTAGATCGTCCTCCT-3' | InDel80 | Chr10-MP7-F | 5'-AGGCGTTGCCTCTAACACTG-3' |
| Chr04-NP8-R | 5'-CAACCGGAGCTGGAAATG-3' | Chr10-MP7-R | 5'-GTCCGGTGAGTTCGTTTCTC-3' | ||
| InDel37 | Chr04-NP11-F | 5'-AGCTCATCAAAGCGAAAGGA-3' | InDel81 | Chr10-MP8-F | 5'-CACATGGTTCTTTTCCACGA-3' |
| Chr04-NP11-R | 5'-AGTCTAGACGCCTCGACGTT-3' | Chr10-MP8-R | 5'-GGAGTTGGATGAAAGCCTTG-3' | ||
| InDel38 | Chr05-MP9-F | 5'-ATGGGAAGGCCTACGATTTT-3' | InDel82 | Chr10-MP11-F | 5'-TGGTAGACGGGTCCTAGTGC-3' |
| Chr05-MP9-R | 5'-TGGGTGATGGTAGGAGAATAGA-3' | Chr10-MP11-R | 5'-TACATCGCTGCTGGAGTGTC-3' | ||
| InDel39 | Chr05-MP13-F | 5'-ATTTAACCTGGCTGCCTGTC-3' | InDel83 | Chr10-NP4-F | 5'-ACACGGGTCGAAGATCAAAG-3' |
| Chr05-MP13-R | 5'-GACCGCGACACTAACAAATG-3' | Chr10-NP4-R | 5'-GGTGCCAACTTCCTTCATCA-3' | ||
| InDel40 | Chr05-MP14-F | 5'-TTTGCCTAGCCATTTCAGGT-3' | InDel84 | Chr10-NP6-F | 5'-TTTGCAAGCTGTTTTGGTTC-3' |
| Chr05-MP14-R | 5'-CACGGTCAACAATCAGCTCT-3' | Chr10-NP6-R | 5'-GGGTGAAGGCTACATCGGTA-3' | ||
| InDel41 | Chr05-NP1-F | 5'-AGCATTGGCTGCTAAGCATT-3' | InDel85 | Chr10-NP7-F | 5'-TACCGTTGTCCGACGCTAAT-3' |
| Chr05-NP1-R | 5'-TAGCAGCAACAGCAAACAGG-3' | Chr10-NP7-R | 5'-TTGTGTAGGCGTCGTGCTTA-3' | ||
| InDel42 | Chr05-NP2-F | 5'-AGAGAGAATCCTCGGCAAGA-3' | InDel86 | Chr10-NP9-F | 5'-CGACCCTGTCGTATGTATGC-3' |
| Chr05-NP2-R | 5'-TTTGTGAGGGGTTCTTGAGC-3' | Chr10-NP9-R | 5'-TTGTCCTTCTGTGGCTGTTG-3' | ||
| InDel43 | Chr05-NP5-F | 5'-CTTCGTGGACCACATGACAC-3' | InDel87 | Chr10-NP11-F | 5'-ACGGAATGCAAAAAGCAAAG-3' |
| Chr05-NP5-R | 5'-GTACCCTCGCCAGTCAACTC-3' | Chr10-NP11-R | 5'-GGTGACTTCAACCGGGATTA-3' | ||
| InDel44 | Chr05-NP6-F | 5'-TCTCACGGGTCGAAAGTACC-3' | |||
| Chr05-NP6-R | 5'-CAATCTCGATTGCCTCAACA-3' |

图1 高粱87个InDel标记PCR产物琼脂糖凝胶电泳检测结果注:M—DNA Marker (D5000); ①—高粱BTx623; ②—高粱JIUTIAN1;1~87分别代表不同InDel标记的PCR产物。
Fig. 1 Agarose-gel electrophoresis detection results of 87 InDel-labeled PCR products in sorghum

图3 利用高粱InDel分子标记进行高粱品种遗传多样性分析的电泳图谱注:M—DNA Marker(D2000),1—白蛇眼,2—Wheatland,3—河北糯杂,4—丽欧,5—T32,6—SEM-E32,7—SEM-42,8—ZIH08894,9—E-Tian,10—JIUTIAN1,11—BTx623。
Fig. 3 Electrophoretic map of genetic diversity analysis of sorghum varieties using InDel markers
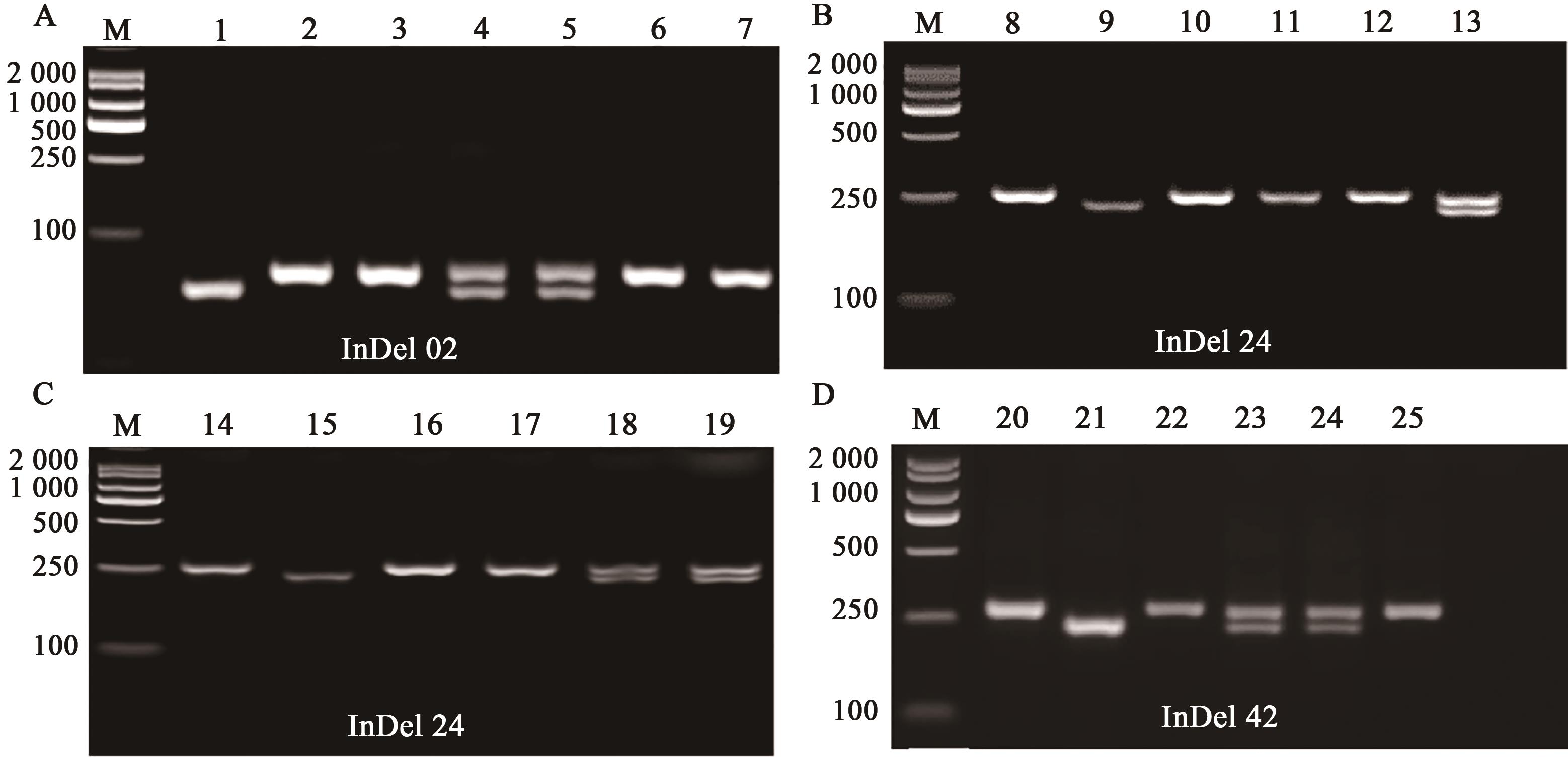
图4 利用InDel标记进行高粱杂交种鉴定的电泳图谱注:M—DNA Marker (D2000) Ⅱ;A~D分别是InDel 02、InDel 24和InDel 42引物的PCR产物电泳结果。1—YM1;2—BTx623;3~7—YM1与 BTx623杂交子代;8—YM2;9—BTx623;10~13—YM2与BTx623杂交子代;14—YM1;15—BTx623;16~19—YM1与BTx623杂交子代;20—YM2;21—BTx623;22~25—YM2与BTx623杂交子代。
Fig. 4 Electrophoretic map of sorghum hybrid identification using InDel markers
| 1 | TARI I, LASKAY G, TAKACS Z, et al.. Response of Sorghum to abiotic stresses: a review[J/OL]. J. Agron. Crop Sci., 2013, 199(4): 12017 [2023-05-23]. . |
| 2 | MCCORMICK R F, TRUONG S K, SREEDASYAM A, et al.. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization[J]. Plant J., 2018, 93(2): 338-354. |
| 3 | LUO H, ZHAO W, WANG Y, et al.. SorGSD: a sorghum genome SNP database[J/OL]. Biotechnol. Biofuels, 2016, 9: 6[2023-05-23]. . |
| 4 | 杨洁, 赫佳, 王丹碧, 等. InDel标记的研究和应用进展[J]. 生物多样性, 2016, 24(2): 237-243.. |
| 5 | VISSCHER P M, YENGO L, COX N J, et al.. Discovery and implications of polygenicity of common diseases[J]. Science, 2021, 373(6562): 1468-1473. |
| 6 | 王钰, 马卉, 许学, 等. 水稻功能性插入缺失标记(InDel)的筛选与应用[J]. 作物杂志, 2019(4): 84-93. |
| 7 | 徐鹏, 蔡继鸿, 杨阳, 等. 陆地棉耐盐相关EST-SSR以及EST-InDel分子标记的开发[J]. 棉花学报, 2016, 28(1): 65-74. |
| 8 | 毛艇, 李旭, 李振宇, 等. 水稻Wx复等位基因基于PCR的功能标记开发与利用[J]. 作物学报, 2017, 43(11): 1715-1723. |
| 9 | YONEMARU J I, ANDO T, MIZUBAYASHI T, et al.. Development of genome-wide simple sequence repeat markers using whole-genome shotgun sequences of Sorghum (Sorghum bicolor (L.) Moench)[J]. DNA Res., 2009, 16(3): 187-193. |
| 10 | 王平, 王春语, 张丽霞, 等. 利用重测序技术开发高粱多态性SSR分子标记[J]. 生物技术通报, 2019, 35(11): 217-223. |
| 11 | BURRELL M, SHARMA A, PATIL N Y, et al.. Sequencing of an anthracnose-resistant Sorghum genotype and mapping of a major QTL reveal strong candidate genes for anthracnose resistance[J]. Crop Sci., 2014, 55(2): 790-799. |
| 12 | ASSEFA A, SHIMELIS H, TONGOONA P, et al. Genetic diversity of lowland sorghum landraces assessed by morphological and microsatellite markers[J]. Aust. J. Crop Sci., 2016, 10(3): 291-298. |
| 13 | BOUCHET S, OLATOYE M O, MARLA S R, et al.. Increased power to dissect adaptive traits in global Sorghum diversity using a nested association mapping population[J]. Genetics, 2017, 206(2): 573-585. |
| 14 | JI G, ZHANG Q, DU R, et al.. Construction of a high-density genetic map using specific-locus amplified fragments in sorghum[J/OL]. BMC Genomics, 2017, 18(1): 51[2023-05-23]. . |
| 15 | CHOE M E, KIM J Y, NABI R B S, et al.. Development of InDels markers for the identification of cytoplasmic male sterility in Sorghum by complete chloroplast genome sequences analysis[J/OL]. Front. Plant Sci., 2023, 14: 1188149[2023-05-23]. . |
| 16 | BOLGER A M, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120. |
| 17 | LI H, DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760. |
| 18 | LALITHA S. Primer Premier 5[R]. Biotech Software & Internet Report, 2000, 270-272. |
| 19 | 李荣华, 夏岩石, 刘顺枝, 等. 改进的CTAB提取植物DNA方法[J]. 实验室研究与探索, 2009, 28(9): 14-16. |
| 20 | VOORRIPS R E. MapChart: software for the graphical presentation of linkage maps and QTLs[J]. J. Hered., 2002, 93(1): 77-78. |
| 21 | 赵文杰, 徐薇, 张景龙, 等. 甜高粱生物学性状与SSR分子标记遗传多样性[J]. 浙江农业学报, 2019, 31(12): 1945-1954. |
| 22 | HAN Y, LV P, HOU S, et al.. Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in Sorghum (Sorghum bicolor L. moench)[J/OL]. PLoS ONE, 2015, 10(5): e0127065[2023-05-23]. . |
| 23 | 胡陶铸, 林丽, 胡继军, 等. 利用InDel标记构建番茄新品种指纹图谱[J]. 上海交通大学学报(农业科学版), 2019, 37(2): 24-29. |
| 24 | 吴迷, 汪念, 沈超, 等. 基于重测序的陆地棉InDel标记开发与评价[J]. 作物学报, 2019, 45(2): 196-203. |
| 25 | 杨易, 黎庭耀, 李国景, 等. 基于重测序的长豇豆基因组InDel标记开发及应用[J]. 园艺学报, 2022, 49(4): 778-790. |
| 26 | TAO Y, LUO H, XU J, et al.. Extensive variation within the pan-genome of cultivated and wild sorghum[J]. Nat. Plants, 2021, 7(6): 766-773. |
| 27 | LI J, WANG L, ZHAN Q, et al.. Transcriptome characterization and functional marker development in Sorghum sudanense[J/OL]. PLoS ONE, 2016, 11(5): e0154947[2023-05-23]. . |
| 28 | BOATWRIGHT J L, SAPKOTA S, KRESOVICH S. Functional genomic effects of indels using Bayesian genome-phenome wide association studies in sorghum[J/OL]. Front. Genet., 2023, 14: 1143395[2023-05-23]. . |
| [1] | 陈峰,徐建第,姜明松,梁水美,张全芳,王文良,李广贤,杨连群,朱文银,周学标. 黄淮区粳稻抗稻瘟病基因Pi-ta、Pi-b、Pi54、Pikm的分子检测 [J]. 生物技术进展, 2018, 8(1): 46-54. |
| [2] | 刘伟,蒋侬辉,袁沛元,邱燕萍,凡超,杨晓燕,向旭. SSR和SNP标记在荔枝遗传育种中的应用[J]. 生物技术进展, 2017, 7(1): 7-12. |
| [3] | 张正,连灵燕,王高鸿,杜艳伟,李颜方,赵晋锋. 分子标记技术在玉米育种中的应用[J]. 生物技术进展, 2015, 5(4): 259-264. |
| [4] | 宋志红,孟庆忠,张涛,张胜昔,王贵春,李国荣. ISSR分子标记在棉花遗传育种上的应用[J]. 生物技术进展, 2014, 4(6): 411-414. |
| [5] | 朱兴彪,郑玉忠,谢丽玲,张振霞,查广才,张勇,刘亚群. 有害赤潮藻种塔玛亚历山大藻的研究进展[J]. 生物技术进展, 2014, 4(6): 421-428. |
| [6] | 何虎翼,谭冠宁,何新民,何海旺,李丽淑,唐洲萍,王晖. 几种基于PCR的分子标记在甘薯遗传多样性研究中的应用[J]. 生物技术进展, 2014, 4(4): 245-250. |
| [7] | 吴星波,郝俊杰,张晓艳,万述伟,李红卫,邵阳,孙吉禄. 普通菜豆抗白粉病基因SCAR标记鉴定[J]. 生物技术进展, 2013, 3(5): 357-362. |
| [8] | 唐三元,席在星,谢旗. 甜高粱在生物能源产业发展中的前景[J]. 生物技术进展, 2012, 2(2): 81-86. |
| [9] | 李长生,宋淑玉,张文刚,夏晗,赵传志. 花生EST数据库的分析、评价和应用前景[J]. 生物技术进展, 2011, 1(3): 172-177. |
| [10] | 王寒玉,杜艳伟,李萍,张喜文,朱晶莹,赵晋锋,余爱丽. 40份玉米自交系的ISSR遗传多样性分析[J]. 生物技术进展, 2011, 1(3): 214-218. |
| [11] | 童继平,韩傲男,王胜军,刘学军,苏京平. 作物数量性状研究进展[J]. 生物技术进展, 2011, 1(2): 98-104. |
| [12] | 罗黎明,刘丽,于丽娟,番兴明. DNA分子标记技术在玉米种子纯度鉴定中的应用[J]. 生物技术进展, 2011, 1(1): 7-13. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部