


生物技术进展 ›› 2022, Vol. 12 ›› Issue (6): 825-836.DOI: 10.19586/j.2095-2341.2022.0048
收稿日期:2022-04-02
接受日期:2022-06-30
出版日期:2022-11-25
发布日期:2022-11-30
通讯作者:
郭忠建
作者简介:周水娟 E-mail: 2807948441@qq.com;
基金资助:
Shuijuan ZHOU( ), Zhongjian GUO(
), Zhongjian GUO( )
)
Received:2022-04-02
Accepted:2022-06-30
Online:2022-11-25
Published:2022-11-30
Contact:
Zhongjian GUO
摘要:
病毒侵染宿主的过程存在着一系列相互作用,了解病毒与宿主之间的蛋白质相互作用对于深入研究病毒具有重要意义。在众多研究蛋白质相互作用的方法中,双分子荧光互补技术(bimolecular fluorescence complementation,BiFC)因其能在活细胞中可视化相互作用而被广泛应用。介绍了双分子荧光技术的原理、发展和优势,总结了双分子荧光技术在动物病毒以及抗病毒药物研究中的应用,并进一步阐述了新型双分子荧光系统的原理,以期为研究动物病毒致病机制和抗病毒药物研发提供新的思路。
中图分类号:
周水娟, 郭忠建. 双分子荧光互补技术在动物病毒中的研究进展[J]. 生物技术进展, 2022, 12(6): 825-836.
Shuijuan ZHOU, Zhongjian GUO. Progress of Bimolecular Fluorescence Complementation Technology in Animal Viruses Research[J]. Current Biotechnology, 2022, 12(6): 825-836.
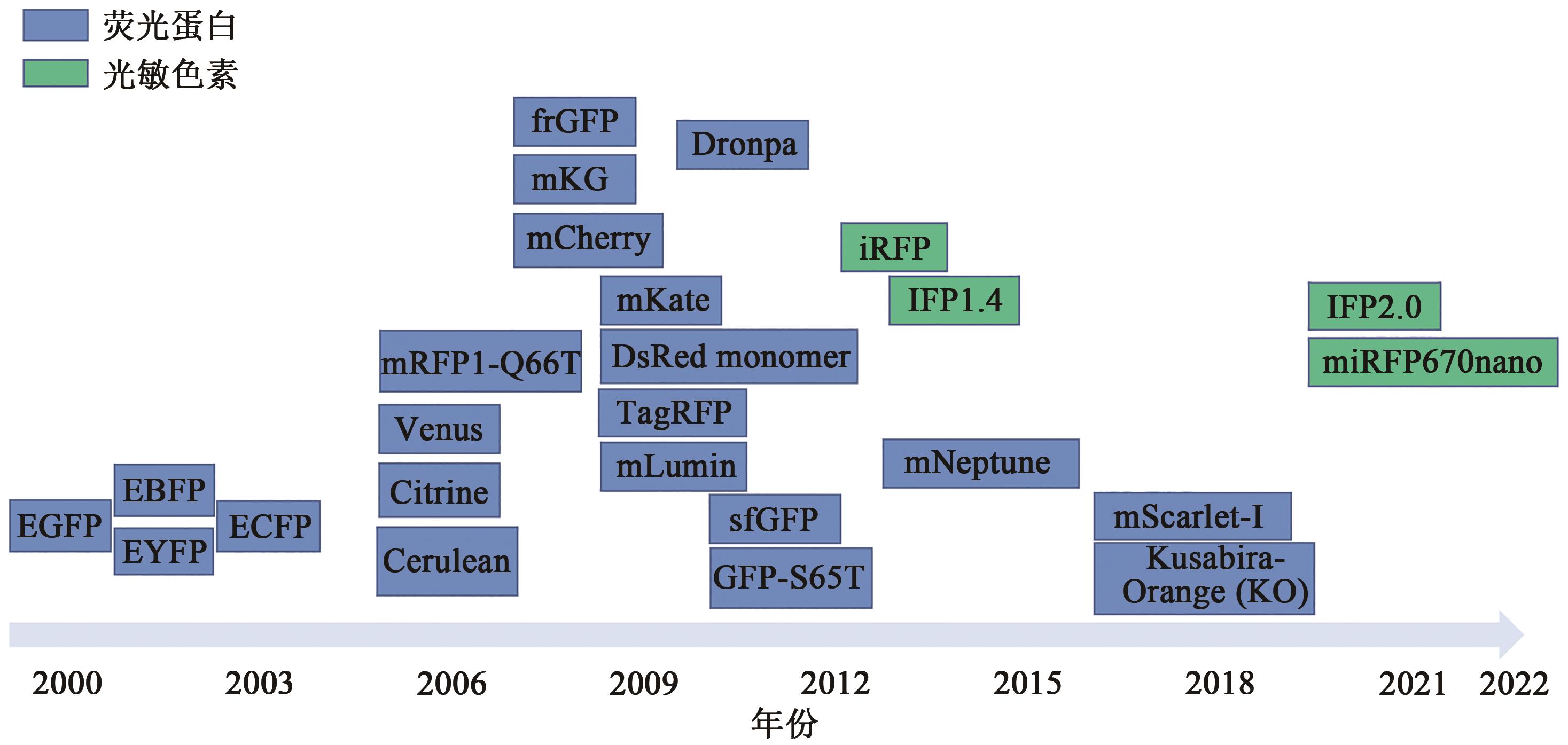
图2 BiFC技术中荧光蛋白和光敏色素发展注:EGFP—增强型绿色荧光蛋白;EBFP—增强型蓝色荧光蛋白;EYFP—增强型黄色荧光蛋白;ECFP—增强型青色荧光蛋白;mRFP1-Q66T—单体红色荧光蛋白变体(Q66T);Venus—黄色荧光蛋白变体(F46L/F64L/M153T/V163A/S175G);Citrine—黄色荧光蛋白变体(Q69M);Cerulean—青色荧光蛋白变体(S72A/Y145A/H148D);frGFP—折叠报告器GFP;mKG—单体珊瑚荧光报告蛋白;mCherry—樱桃色荧光蛋白;mKate—远红荧光蛋白报告基因;DsRed monomer—红色荧光蛋白单体;TagRFP—橘红色荧光蛋白;mLumin—mKate-S158A变体;Dronpa—单体GFP样荧光蛋白;sfGFP—一种设计精良的折叠版本GFP突变体(S30R/Y39N/N105T/Y145F/I171V/A206V);GFP-S65T—绿色荧光蛋白突变体(S65T);iRFP—近红外荧光蛋白;IRF1.4—近红外荧光蛋白1.4;mNeptune—mKate变体(M41G/S61C/S158C/Y194F/);mScarlet-I—单体红色荧光蛋白变体;Kusabira-Orange(KO)—橙色荧光蛋白;IFP2.0—近红外荧光蛋白2.0;miRFP670namo—最小的近红外荧光蛋白。
Fig. 2 The development of fluorescent proteins and photochromes in BiFC
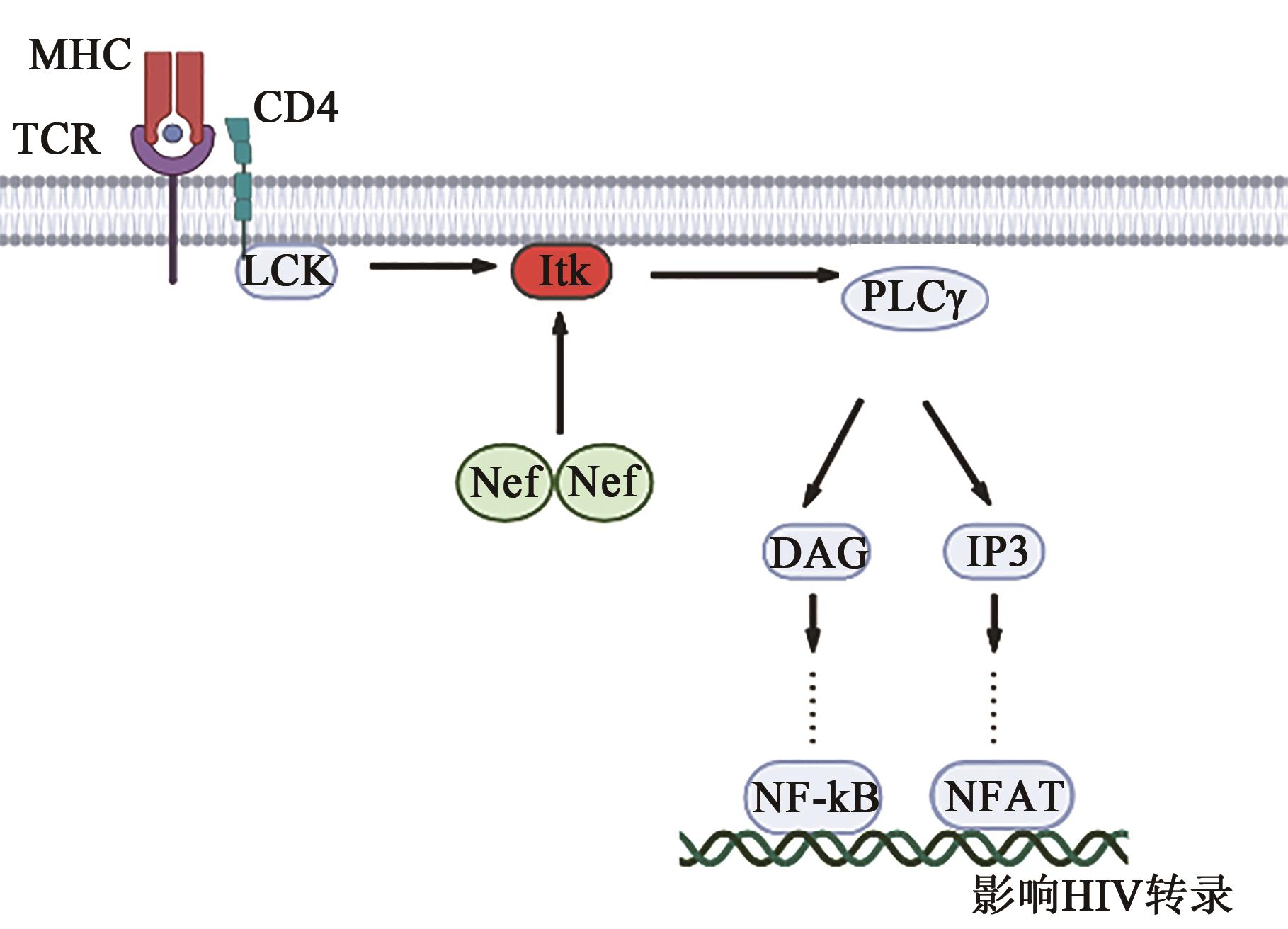
图3 Nef二聚体与Itk相互作用激活模型[37]注:MHC—组织相容性复合物;TCR—T细胞抗原受体;CD4—表面抗原分化簇4受体;LCK—蛋白酪氨酸激酶;PLC γ—磷脂酶C γ链;DAG—二酰基甘油;IP3—三磷酸肌醇;NF-κB—核转录因子;NFAT—活化T细胞核因子。
Fig. 3 Models of Nef-dimer mediated Itk activation[37]
| 病毒种类 | 相互作用的蛋白 | 荧光标签(荧光蛋白和光敏色素) | 相互作用定位 | 参考文献 |
|---|---|---|---|---|
| 人类免疫缺陷病毒 | IN与importin α | GFP | 细胞核 | [ |
| IN与LEDGF/p75 | iRFP | 染色体 | [ | |
| IN多聚体 | Venus | 细胞质 | [ | |
| Nef二聚体 | YFP | 细胞膜和高尔基体网络 | [ | |
| Nef与Itk、Btk | Venus | 细胞膜 | [ | |
| Nef与AP-2、SERINC5 | Venus | 细胞膜 | [ | |
| Nef与PACS | YFP | 细胞质 | [ | |
| Nef与SNX18 | Venus | 细胞质 | [ | |
| Nef与Tim-3 | Venus | 细胞质 | [ | |
| Env与Env、SERINC5 | Venus | 细胞膜和细胞质 | [ | |
| Vpu与Tim-3 | Venus | 未知 | [ | |
| Gag与Gag、AGO2 | Venus | 未知 | [ | |
| 疱疹病毒 | gD与gB、gH/gL | Venus | 细胞质和细胞核 | [ |
| gB与gH/gL | Venus | 细胞质和细胞核 | [ | |
| ICP27寡聚体 | Venus | 细胞核 | [ | |
| ICP27与TAP/NXF1 | Venus | 细胞核 | [ | |
| HSRG1与LT | YFP | 细胞核 | [ | |
| LMP1与LMP1、TRAF2、TRAF3 | YFP | 细胞膜及核周 | [ | |
| LMP1与Tmem134 | YFP | 细胞质 | [ | |
| LMP1与肌动蛋白细胞骨架相关蛋白 | YFP | 脂筏 | [ | |
| pUL21与Roadblock-1 | Lumin | 细胞质 | [ | |
| pUL14与VP16 | Venus | 细胞核 | [ | |
| 流感病毒 | PA与PB1 | Venus | 细胞核 | [ |
| PB1与PB2 | Venus | 细胞核 | [ | |
| PA与PB2 | Venus | 细胞核 | [ | |
| RIG I与PB2 | Venus | 细胞核 | [ | |
| RIG I与PA、PB1 | Venus | 细胞质 | [ | |
| 其他病毒 | NS4B与STING | mKG | 细胞质 | [ |
| E2与Brd4 | Venus | 细胞核 | [ | |
| VP2与Apoptin | YFP | 细胞核 | [ | |
| RIG-I与TRIM25、MAVS | Venus | 细胞质 | [ | |
| PRRSV跨膜非结构蛋白互作网络 | Venus | 未知 | [ | |
| poMx1与NS5B | Venus | 细胞质 | [ | |
| BmCdc37与BmHsp90 | Venus | 细胞质 | [ | |
| Claudin-2与VP7 | dsRed | 未知 | [ | |
| SARS-CoV-2结构蛋白和辅助蛋白 | Venus | 未知 | [ | |
| SARS-CoV-2核衣壳蛋白与细胞应激颗粒蛋白 | miRFP670nano | 细胞质 | [ |
表1 BiFC在致病性病毒蛋白互作研究中的应用
Table 1 Application of BiFC in the study of proteins interaction of pathogenic viruses
| 病毒种类 | 相互作用的蛋白 | 荧光标签(荧光蛋白和光敏色素) | 相互作用定位 | 参考文献 |
|---|---|---|---|---|
| 人类免疫缺陷病毒 | IN与importin α | GFP | 细胞核 | [ |
| IN与LEDGF/p75 | iRFP | 染色体 | [ | |
| IN多聚体 | Venus | 细胞质 | [ | |
| Nef二聚体 | YFP | 细胞膜和高尔基体网络 | [ | |
| Nef与Itk、Btk | Venus | 细胞膜 | [ | |
| Nef与AP-2、SERINC5 | Venus | 细胞膜 | [ | |
| Nef与PACS | YFP | 细胞质 | [ | |
| Nef与SNX18 | Venus | 细胞质 | [ | |
| Nef与Tim-3 | Venus | 细胞质 | [ | |
| Env与Env、SERINC5 | Venus | 细胞膜和细胞质 | [ | |
| Vpu与Tim-3 | Venus | 未知 | [ | |
| Gag与Gag、AGO2 | Venus | 未知 | [ | |
| 疱疹病毒 | gD与gB、gH/gL | Venus | 细胞质和细胞核 | [ |
| gB与gH/gL | Venus | 细胞质和细胞核 | [ | |
| ICP27寡聚体 | Venus | 细胞核 | [ | |
| ICP27与TAP/NXF1 | Venus | 细胞核 | [ | |
| HSRG1与LT | YFP | 细胞核 | [ | |
| LMP1与LMP1、TRAF2、TRAF3 | YFP | 细胞膜及核周 | [ | |
| LMP1与Tmem134 | YFP | 细胞质 | [ | |
| LMP1与肌动蛋白细胞骨架相关蛋白 | YFP | 脂筏 | [ | |
| pUL21与Roadblock-1 | Lumin | 细胞质 | [ | |
| pUL14与VP16 | Venus | 细胞核 | [ | |
| 流感病毒 | PA与PB1 | Venus | 细胞核 | [ |
| PB1与PB2 | Venus | 细胞核 | [ | |
| PA与PB2 | Venus | 细胞核 | [ | |
| RIG I与PB2 | Venus | 细胞核 | [ | |
| RIG I与PA、PB1 | Venus | 细胞质 | [ | |
| 其他病毒 | NS4B与STING | mKG | 细胞质 | [ |
| E2与Brd4 | Venus | 细胞核 | [ | |
| VP2与Apoptin | YFP | 细胞核 | [ | |
| RIG-I与TRIM25、MAVS | Venus | 细胞质 | [ | |
| PRRSV跨膜非结构蛋白互作网络 | Venus | 未知 | [ | |
| poMx1与NS5B | Venus | 细胞质 | [ | |
| BmCdc37与BmHsp90 | Venus | 细胞质 | [ | |
| Claudin-2与VP7 | dsRed | 未知 | [ | |
| SARS-CoV-2结构蛋白和辅助蛋白 | Venus | 未知 | [ | |
| SARS-CoV-2核衣壳蛋白与细胞应激颗粒蛋白 | miRFP670nano | 细胞质 | [ |
| 1 | KODAMA Y, HU C D. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives[J]. Biotechniques, 2012, 53(5): 285-298. |
| 2 | GHOSH I, HAMILTON A D, REGAN L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein[J]. J. Am. Chem. Soc., 2000, 122(23): 5658-5659. |
| 3 | HU C D, CHINENOV Y, KERPPOLA T K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation[J]. Mol. Cell, 2002, 9(4): 789-798. |
| 4 | HU C D, KERPPOLA T K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis[J]. Nat. Biotechnol., 2003, 21(5): 539-545. |
| 5 | SHYU Y J, LIU H, DENG X, et al.. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions[J]. Biotechniques, 2006, 40(1): 61-66. |
| 6 | SARKAR M, MAGLIERY T J. Re-engineering a split-GFP reassembly screen to examine RING-domain interactions between BARD1 and BRCA1 mutants observed in cancer patients[J]. Mol. Biosyst., 2008, 4(6): 599-605. |
| 7 | YUTAKA K. A bright green-colored bimolecular fluorescence complementation assay in living plant cells[J]. Plant Biotechnol., 2011, 28(1): 95-98. |
| 8 | ZHOU J, LIN J, ZHOU C, et al.. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein[J]. Acta Biochim. Biophys. Sin., 2011, 43(3): 239-244. |
| 9 | UEYAMA T, KUSAKABE T, KARASAWA S, et al.. Sequential binding of cytosolic Phox complex to phagosomes through regulated adaptor proteins: evaluation using the novel monomeric Kusabira-Green system and live imaging of phagocytosis[J]. J. Immunol., 2008, 181(1): 629-640. |
| 10 | LEE Y R, PARK J H, HAHM S H, et al.. Development of bimolecular fluorescence complementation using Dronpa for visualization of protein-protein interactions in cells[J]. Mol. Imaging Biol., 2010, 12(5): 468-478. |
| 11 | JACH G, PESCH M, RICHTER K, et al.. An improved mRFP1 adds red to bimolecular fluorescence complementation[J]. Nat. Methods, 2006, 3(8): 597-600. |
| 12 | FAN J Y, CUI Z Q, WEI H P, et al.. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells[J]. Biochem. Biophys. Res. Commun., 2008, 367(1): 47-53. |
| 13 | CHU J, ZHANG Z, ZHENG Y, et al.. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions[J]. Biosens. Bioelectron., 2009, 25(1): 234-239. |
| 14 | QIN L, CHU J, YING Z, et al.. A new red bimolecular fluorescence complementation based on TagRFP[C]. //Proceedings of SPIE-The International Society for Optical Engineering, 2009, 7191. |
| 15 | HAN Y, WANG S, ZHANG Z, et al.. In vivo imaging of protein-protein and RNA-protein interactions using novel far-red fluorescence complementation systems[J/OL]. Nucl. Acids Res., 2014, 42(13): e103[2014-05-09]. . |
| 16 | WANG S, DING M, XUE B, et al.. Spying on protein interactions in living cells with reconstituted scarlet light[J]. Analyst, 2018, 143(21): 5161-5169. |
| 17 | FUJII Y, YOSHIMURA A, KODAMA Y, et al.. A novel orange-colored bimolecular fluorescence complementation (BiFC) assay using monomeric Kusabira-Orange protein[J]. Biotechniques, 2018, 64(4): 153-161. |
| 18 | FANKHAUSER C. The phytochromes, a family of red/far-red absorbing photoreceptors[J]. J. Biol. Chem., 2001, 276(15): 11453-11456. |
| 19 | FILONOV G S, VERKHUSHA V V. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions[J]. Chem. Biol.,2013, 20(8): 1078-1086. |
| 20 | CHEN M, LI W, ZHANG Z, et al.. Novel near-infrared BiFC systems from a bacterial phytochrome for imaging protein interactions and drug evaluation under physiological conditions[J]. Biomaterials, 2015, 48: 97-107. |
| 21 | TCHEKANDA E, SIVANESAN D, MICHNICK S W. An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions[J]. Nat. Methods, 2014, 11(6): 641-644. |
| 22 | CHEN M, YAN C, MA Y, et al.. A tandem near-infrared fluorescence complementation system with enhanced fluorescence for imaging protein-protein interactions in vivo[J/OL]. Biomaterials, 2021, 268: 120544[2021-11-23]. . |
| 23 | CHEN M, YAN C, ZHENG L, et al.. The smallest near-infrared fluorescence complementation system for imaging protein-protein and RNA-protein interactions[J]. Chem. Sci., 2021, 13(4): 1119-1129. |
| 24 | MAGLIERY T J, WILSON C G, PAN W, et al.. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism[J]. J. Am. Chem. Soc., 2005, 127(1): 146-157. |
| 25 | MORELL M, ESPARGARÓ A, AVILÉS F X, et al.. Detection of transient protein-protein interactions by bimolecular fluorescence complementation: the Abl-SH3 case[J]. Proteomics, 2007, 7(7): 1023-1036. |
| 26 | KERPPOLA T K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells[J]. Annu. Rev. Biophys., 2008, 37: 465-487. |
| 27 | HU C D, KERPPOLA T K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis[J]. Nat. Biotechnol., 2003, 21(5): 539-545. |
| 28 | LEVIN A, ARMON-OMER A, ROSENBLUH J, et al.. Inhibition of HIV-1 integrase nuclear import and replication by a peptide bearing integrase putative nuclear localization signal[J]. Retrovirology, 2009, 6(1):1-16. |
| 29 | CHEN M, LI W, ZHANG Z, et al.. Novel near-infrared BiFC systems from a bacterial phytochrome for imaging protein interactions and drug evaluation under physiological conditions[J]. Biomaterials, 2015, 48: 97-107. |
| 30 | MAERTENS G, CHEREPANOV P, PLUYMERS W, et al.. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells[J]. J. Biol. Chem., 2003, 278(35): 33528-33539. |
| 31 | SHUN M C, RAGHAVENDRA N K, VANDEGRAAFF N, et al.. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration[J]. Genes Dev., 2007, 21(14): 1767-1778. |
| 32 | NAKAMURA T, CAMPBELL J R, MOORE A R, et al.. Development and validation of a cell-based assay system to assess human immunodeficiency virus type 1 integrase multimerization[J]. J. Virol. Methods, 2016, 236: 196-206. |
| 33 | GUIOT E, CARAYON K, DELELIS O, et al.. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity[J]. J. Biol. Chem., 2006, 281(32): 22707-22719. |
| 34 | AROLD S, HOH F, DOMERGUE S, et al.. Characterization and molecular basis of the oligomeric structure of HIV-1 Nef protein[J]. Protein Sci., 2000, 9(6): 1137-1148. |
| 35 | LEE C H, SAKSELA K, MIRZA U A, et al.. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain[J]. Cell, 1996, 85(6): 931-942. |
| 36 | POE J A, SMITHGALL T E. HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication[J]. J. Mol. Biol., 2009, 394(2): 329-342. |
| 37 | LI W F, ARYAL M, SHU S T, et al.. HIV-1 Nef dimers short-circuit immune receptor signaling by activating Tec-family kinases at the host cell membrane[J]. J. Biol. Chem., 2020, 295(15): 5163-5174. |
| 38 | STAUDT R P, SMITHGALL T E. Nef homodimers down-regulate SERINC5 by AP-2-mediated endocytosis to promote HIV-1 infectivity[J]. J. Biol. Chem., 2020, 295(46): 15540-15552. |
| 39 | SHU S T, EMERT-SEDLAK L A, SMITHGALL T E. Cell-based fluorescence complementation reveals a role for HIV-1 Nef protein dimerization in AP-2 adaptor recruitment and CD4 co-receptor down-regulation[J]. J. Biol. Chem., 2017, 292(7): 2670-2678. |
| 40 | DIKEAKOS J D, THOMAS L, KWON G, et al.. An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to down-regulate MHC-I[J]. Mol. Biol. Cell, 2012, 23(11): 2184-2197. |
| 41 | DIRK B S, JACOB R A, JOHNSON A L, et al.. Viral bimolecular fluorescence complementation: a novel tool to study intracellular vesicular trafficking pathways[J/OL]. PLoS ONE, 2015, 10(4): e0125619[2015-04-27]. . |
| 42 | JACOB R A, EDGAR C R, PRÉVOST J, et al.. The HIV-1 accessory protein Nef increases surface expression of the checkpoint receptor Tim-3 in infected CD4+ T cells[J/OL]. J. Biol. Chem., 2021, 297(3): 101042[2021-08-04]. . |
| 43 | ZHANG X, SHI J, QIU X, et al.. CD4 expression and Env conformation are critical for HIV-1 restriction by SERINC5[J/OL]. J. Virol., 2019, 93(14): e00544-19[2019-06-28]. . |
| 44 | PRÉVOST J, EDGAR C R, RICHARD J, et al.. HIV-1 Vpu downregulates Tim-3 from the surface of infected CD4+ T cells[J/OL]. J. Virol., 2020, 94(7): e01999-19[2020-03-17]. . |
| 45 | MAO S, YING Y, MA Z, et al.. A background assessable and correctable bimolecular fluorescence complementation system for nanoscopic single-molecule imaging of intracellular protein-protein interactions[J]. ACS Nano, 2021, 15(9): 14338-14346. |
| 46 | ATANASIU D, WHITBECK J C, CAIRNS T M, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion[J]. Proc. Natl. Acad. Sci. USA, 2007, 104(47): 18718-18723. |
| 47 | AVITABILE E, FORGHIERI C, CAMPADELLI-FIUME G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein[J]. J. Virol., 2007, 81(20): 11532-11537. |
| 48 | HERNANDEZ F P, SANDRI-GOLDIN R M. Herpes simplex virus 1 regulatory protein ICP27 undergoes a head-to-tail intramolecular interaction[J]. J. Virol., 2010, 84(9): 4124-4135. |
| 49 | HERNANDEZ F P, SANDRI-GOLDIN R M. Bimolecular fluorescence complementation analysis to reveal protein interactions in herpes virus infected cells[J]. Methods, 2011, 55(2): 182-187. |
| 50 | HERNANDEZ F P, SANDRI-GOLDIN R M. Head-to-tail intramolecular interaction of herpes simplex virus type 1 regulatory protein ICP27 is important for its interaction with cellular mRNA export receptor TAP/NXF1[J/OL]. MBio, 2010, 1(5): e00268-10 [2010-11-09]. . |
| 51 | GUO H X, CUN W, LIU L D, et al.. Protein encoded by HSV-1 stimulation-related gene 1 (HSRG1) interacts with and inhibits SV40 large T antigen[J]. Cell Prolif., 2006, 39(6): 507-518. |
| 52 | YOUNG L S, RICKINSON A B. Epstein-Barr virus: 40 years on[J]. Nat. Rev. Cancer, 2004, 4(10): 757-768. |
| 53 | TALATY P, EMERY A, EVERLY D N. Characterization of the latent membrane protein 1 signaling complex of Epstein-Barr virus in the membrane of mammalian cells with bimolecular fluorescence complementation[J/OL]. Virol. J., 2011, 8: 414[2011-08-24]. . |
| 54 | ALATY P, EMERY A, HOLTHUSEN K, et al.. Identification of transmembrane protein 134 as a novel LMP1-binding protein by using bimolecular fluorescence complementation and an enhanced retroviral mutagen[J]. J. Virol., 2012, 86(20): 11345-11355. |
| 55 | HOLTHUSEN K, TALATY P, EVERLY D N. Regulation of latent membrane protein 1 signaling through interaction with cytoskeletal proteins[J]. J. Virol., 2015, 89(14): 7277-7290. |
| 56 | YAN K, LIU J, GUAN X, et al.. The carboxyl terminus of tegument protein pUL21 contributes to pseudorabies virus neuroinvasion[J/OL]. J. Virol., 2019, 93(7): e02052-18[2019-03-21]. . |
| 57 | LI F, ZHANG Y, CHEN S, et al.. Identification of the nuclear localization signal region of duck enteritis virus UL14 and its interaction with VP16[J]. Intervirology, 2016, 59(4): 187-196. |
| 58 | CHEN L, NI Z, HUA J, et al.. Proteomic analysis of host cellular proteins co-immunoprecipitated with duck enteritis virus gC[J/OL]. J. Proteomics, 2021, 245: 104281[2021-08-15]. . |
| 59 | DENG T, SHARPS J, FODOR E, et al.. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex[J]. J. Virol., 2005, 79(13): 8669-8674. |
| 60 | HEMERKA J N, WANG D, WENG Y, et al.. Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay[J]. J. Virol., 2009, 83(8): 3944-3955. |
| 61 | SUZUKI T, AINAI A, NAGATA N, et al.. A novel function of the N-terminal domain of PA in assembly of influenza A virus RNA polymerase[J]. Biochem. Biophys. Res. Commun., 2011, 414(4): 719-726. |
| 62 | LI W, CHEN H, SUTTON T, et al.. Interactions between the influenza A virus RNA polymerase components and retinoic acid-inducible gene I[J]. J. Virol., 2014, 88(18): 10432-10447. |
| 63 | NITTA S, SAKAMOTO N, NAKAGAWA M, et al.. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity[J]. Hepatology, 2013, 57(1): 46-58. |
| 64 | HELFER C M, WANG R, YOU J. Analysis of the papillomavirus E2 and bromodomain protein Brd4 interaction using bimolecular fluorescence complementation[J/OL]. PLoS ONE, 2013, 8(10): e77994[2013-10-25]. . |
| 65 | SÁNCHEZ-APARICIO M T, AYLLÓN J, LEO-MACIAS A, et al.. Subcellular localizations of RIG-I, TRIM 25, and MAVS complexes[J/OL]. J. Virol., 2017, 91(2): e01155-16[2017-01-03]. . |
| 66 | LAI G H, LIEN Y Y, LIN M K, et al.. VP2 of chicken anaemia virus interacts with apoptin for down-regulation of apoptosis through de-phosphorylated threonine 108 on apoptin[J/OL]. Sci. Rep., 2017, 7(1): 14799[2017-11-01]. . |
| 67 | NAN H, LAN J, TIAN M, et al. The network of interactions among porcine reproductive and respiratory syndrome virus non-structural proteins[J/OL]. Front. Microbiol., 2018, 9: 970[2018-05-14]. . |
| 68 | ZHOU J, CHEN J, ZHANG X M, et al.. Porcine Mx1 protein inhibits classical swine fever virus replication by targeting nonstructural protein NS5B[J/OL]. J. Virol., 2018, 92(7): e02147-17[2022-06-30].. |
| 69 | ZHANG Q, WU Y F, CHEN P, et al.. Bombyx mori cell division cycle protein 37 promotes the proliferation of BmNPV[J/OL]. Pestic. Biochem. Physiol., 2021, 178: 104923[2021-07-17].. |
| 70 | ZHU M, ZHANG X, PAN J, et al.. Tight junction protein claudin-2 promotes cell entry of Bombyx mori cypovirus[J]. Appl. Microbiol. Biotechnol., 2021, 105(14-15): 6019-6031. |
| 71 | CHEN M, YAN C, QIN F, et al.. The intraviral protein-protein interaction of SARS-CoV-2 reveals the key role of N protein in virus-like particle assembly[J]. Int. J. Biol. Sci., 2021, 17(14): 3889-3897. |
| 72 | EMERT-SEDLAK L A, NARUTE P, SHU S T, et al.. Effector kinase coupling enables high-throughput screens for direct HIV-1 Nef antagonists with antiretroviral activity[J]. Chem. Biol., 2013, 20(1): 82-91. |
| 73 | SHUN M C, RAGHAVENDRA N K, VANDEGRAAFF N, et al.. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration[J]. Genes Dev., 2007, 21(14): 1767-1778. |
| 74 | ZHOU Z, JIANG X, LIU D, et al.. Autophagy is involved in influenza A virus replication[J]. Autophagy, 2009, 5(3): 321-328. |
| 75 | DAI J P, LI W Z, ZHAO X F, et al.. A drug screening method based on the autophagy pathway and studies of the mechanism of evodiamine against influenza A virus[J/OL]. PLoS ONE, 2012, 7(8): e42706[2012-08-10].. |
| 76 | MÜNZ C. Beclin-1 targeting for viral immune escape[J]. Viruses, 2011, 3(7): 1166-1178. |
| 77 | DAI J P, ZHAO X F, ZENG J, et al.. Drug screening for autophagy inhibitors based on the dissociation of Beclin1-Bcl2 complex using BiFC technique and mechanism of eugenol on anti-influenza a virus activity[J/OL]. PLoS ONE, 2013, 8(4): e61026[2013-04-16]. . |
| 78 | DAI J, WANG G, LI W, et al.. High-throughput screening for anti-influenza A virus drugs and study of the mechanism of procyanidin on influenza A virus-induced autophagy[J]. J. Biomol. Screen., 2012, 17(5): 605-617. |
| 79 | YU C, LI S, ZHANG X, et al.. MARCH8 inhibits Ebola virus glycoprotein, human immunodeficiency virus type 1 envelope glycoprotein, and avian influenza virus H 5N1 hemagglutinin maturation[J/OL]. mBio, 2020, 11(5): e01882-20[2022-06-30].. |
| 80 | WEI J, HAMEED M, WANG X, et al.. Antiviral activity of phage display-selected peptides against Japanese encephalitis virus infection in vitro and in vivo[J/OL]. Antiviral Res., 2020, 174:104673[2020-12-05]. . |
| 81 | HUANG C, BERNARD D, ZHU J, et al.. Small molecules block the interaction between porcine reproductive and respiratory syndrome virus and CD163 receptor and the infection of pig cells [J/OL]. Virol. J., 2020, 17(1): 116[2020-07-30]. . |
| 82 | WANG Y, WANG Z, LIU J, et al.. Discovery of novel HBV capsid assembly modulators by structure-based virtual screening and bioassays[J/OL]. Bioorg. Med. Chem., 2021, 36(11): 116096[2021-04-15]. . |
| 83 | SHAO S, ZHANG H, ZENG Y, et al.. TagBiFC technique allows long-term single-molecule tracking of protein-protein interactions in living cells[J/OL]. Commun. Biol.. 2021, 4(1): 378[2021-03-19]. . |
| 84 | ZAMYATNIN A A, SOLOVYEV A G, BOZHKOV P V, et al.. Assessment of the integral membrane protein topology in living cells[J]. Plant J., 2006, 46(1): 145-154. |
| 85 | LEASTRO M O, FREITAS-ASTÚA J, KITAJIMA E W, et al.. Dichorhaviruses movement protein and nucleoprotein form a protein complex that may be required for virus spread and interacts in vivo with viral movement-related cilevirus proteins[J/OL]. Front. Microbiol., 2020, 11: 571807[2020-11-04]. . |
| 86 | HAN Y, WANG S, ZHANG Z, et al.. In vivo imaging of protein-protein and RNA-protein interactions using novel far-red fluorescence complementation systems[J/OL]. Nucl. Acids Res., 2014, 42(13): e103[2014-05-09]. . |
| 87 | PARK S Y, MOON H C, PARK H Y. Live-cell imaging of single mRNA dynamics using split superfolder green fluorescent proteins with minimal background[J]. RNA, 2020, 26(1): 101-109. |
| 88 | 熊冰钰,叶玙璠,崔凌云,等.蛋白质-RNA相互作用鉴定技术研究进展[J].生物技术进展, 2020, 10(3): 217-225. |
| 89 | MORELL M, ESPARGARO A, AVILES F X, et al.. Study and selection of in vivo protein interactions by coupling bimolecular fluorescence complementation and flow cytometry[J]. Nat. Protoc., 2008, 3(1): 22-33. |
| 90 | SHYU Y J, SUAREZ C D, HU C D. Visualization of ternary complexes in living cells by using a BiFC-based FRET assay[J]. Nat. Protoc., 2008, 3(11): 1693-1702. |
| [1] | 王芬, 潘发莲, 杨秀, 韦永欢, 裴会敏. 福鼎大白茶蛋白质相互作用网络研究[J]. 生物技术进展, 2022, 12(4): 623-629. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部