


生物技术进展 ›› 2022, Vol. 12 ›› Issue (2): 176-188.DOI: 10.19586/j.2095-2341.2021.0050
收稿日期:2021-04-14
接受日期:2021-12-06
出版日期:2022-03-25
发布日期:2022-03-28
通讯作者:
梁泉峰
作者简介:靳鑫 E‑mail:934183327@qq.com;
基金资助:
Xin JIN( ), Sumeng WANG, Qingsheng QI, Quanfeng LIANG(
), Sumeng WANG, Qingsheng QI, Quanfeng LIANG( )
)
Received:2021-04-14
Accepted:2021-12-06
Online:2022-03-25
Published:2022-03-28
Contact:
Quanfeng LIANG
摘要:
L?异亮氨酸属于三大支链氨基酸,是人体8种必需氨基酸之一,广泛应用于食品、药品、保健品、化妆品等领域。目前,微生物发酵法是工业生产L?异亮氨酸的主要方法,其中谷氨酸棒杆菌(Corynebacterium glutamicum)是发酵生产L?异亮氨酸的优势菌株,然而随机诱变会使产量的提高能力达到饱和,难以得到更加高产的菌株,因此针对诱变菌株进行理性改造已成为进一步提高产量的主要方式;且随着遗传操作技术在谷氨酸棒杆菌中的应用与优化,代谢工程育种已逐渐取代传统的诱变育种。综述了谷氨酸棒杆菌中L?异亮氨酸的生物合成途径、代谢调控机制和理性改造L?异亮氨酸生产菌株的策略,并对辅助因子工程应用于理性改造及对谷氨酸棒杆菌基因组整合策略进行了系统阐述,以期为工业水平稳定生产L?异亮氨酸高产菌株的基因组整合策略提供参考依据。
中图分类号:
靳鑫, 王苏蒙, 祁庆生, 梁泉峰. 谷氨酸棒杆菌生产异亮氨酸辅因子策略及其基因组整合研究进展[J]. 生物技术进展, 2022, 12(2): 176-188.
Xin JIN, Sumeng WANG, Qingsheng QI, Quanfeng LIANG. Cofactor Strategy and Genome Integration of L‑isoleucine Production by Corynebacterium glutamicum[J]. Current Biotechnology, 2022, 12(2): 176-188.
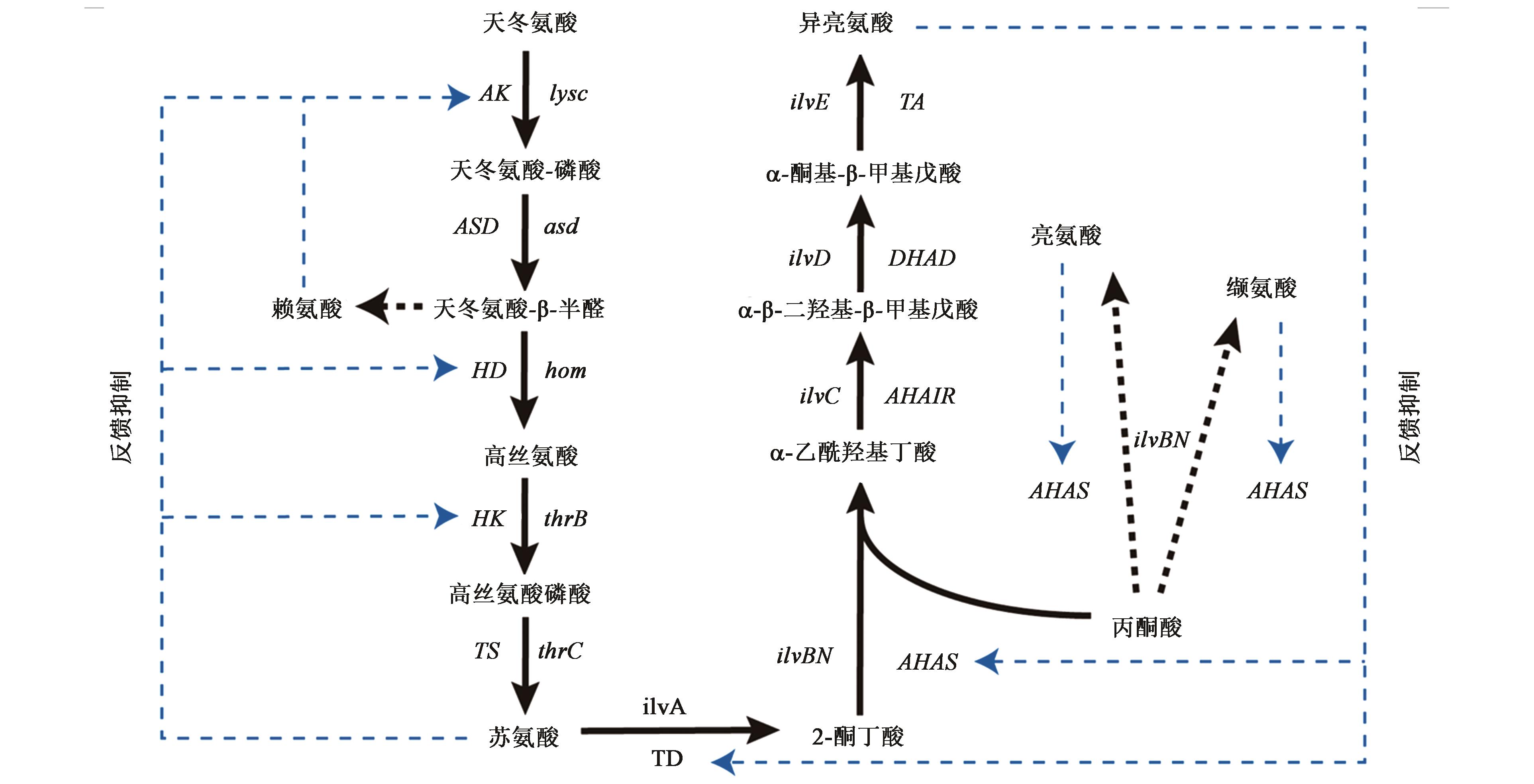
图1 谷氨酸棒杆菌中L?异亮氨酸的生物合成途径及代谢调控机制注:AK—天冬氨酸激酶;ASD—天冬氨酸半醛脱氢酶;HD—高丝氨酸脱氢酶;HK—高丝氨酸激酶;TS—苏氨酸合成酶;TD:苏氨酸脱水酶;AHAS—乙酰羟基酸合成酶;AHAIR—乙酰羟酸异构还原酶;DHAD—二羟酸还原异构酶;TA—支链氨基酸转氨酶
Fig.1 Biosynthetic pathway and metabolic regulation mechanism of L?isoleucine in Corynebacterium glutamicum
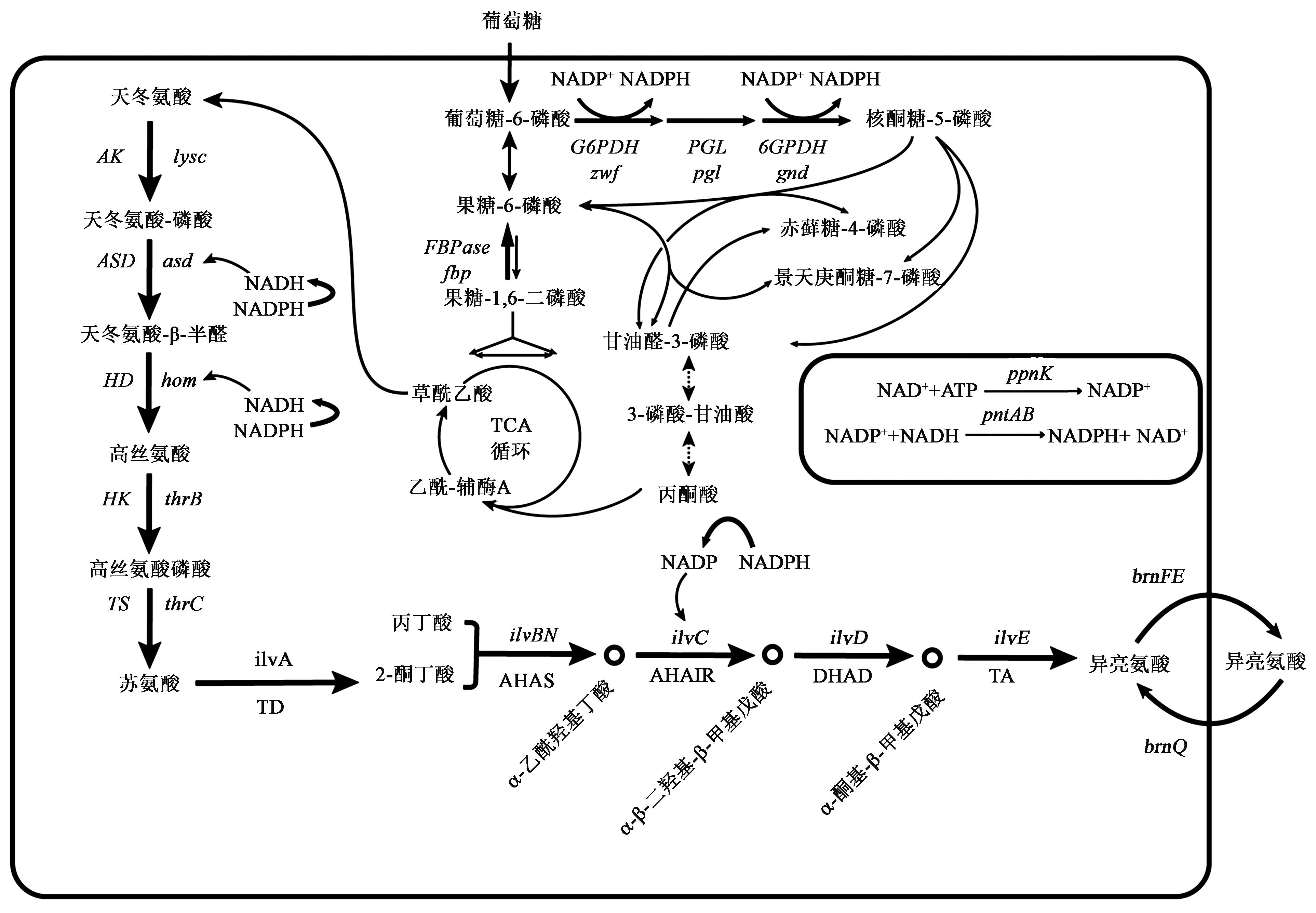
图2 谷氨酸棒杆菌中L?异亮氨酸的改造策略注:AK—天冬氨酸激酶;ASD—天冬氨酸半醛脱氢酶;HD—高丝氨酸脱氢酶;HK—高丝氨酸激酶;TS—苏氨酸合成酶;TD:苏氨酸脱水酶;AHAS—乙酰羟基酸合成酶;AHAIR—乙酰羟酸异构还原酶;DHAD—二羟酸还原异构酶;TA—支链氨基酸转氨酶。NADH—还原型辅酶Ⅰ;NADP+—烟酰胺腺嘌呤二核苷酸;NADPH—还原型辅酶Ⅱ;brnFE—编码双组分转运系统;brnQ—编码L?异亮氨酸胞内转运;zwf—编码葡萄糖?6?磷酸脱氢酶(G6PDH);pgl—编码6?磷酸葡萄糖酸内酯酶(PGL);gnd—编码6?磷酸葡萄糖酸脱氢酶(6GPDH);fbp—编码果糖?1,6?磷酸酶(FBPase);ppnK—NAD+激酶编码基因;pntAB—膜结合转氢酶编码基因。
Fig.2 Transformation strategy of L?isoleucine in Corynebacterium glutamicum
| 受底物反馈抑制的酶 | 底物 | 解除反馈抑制位点 | 参考文献 |
|---|---|---|---|
| 天冬氨酸激酶 | 苏氨酸 | N374A、D274A、E278V、G277A、A279V | [ |
| 赖氨酸 | T361A、S381F | [ | |
| 苏氨酸、赖氨酸 | N299L、T336L、M365A、N372A、I272E、Q298G、I375P、E382A、F283R、R384L、S386A、S301F、T308A、F364A、T311I、A279T | [ | |
| 高丝氨酸脱氢酶 | 苏氨酸 | G378E、G378S | [ |
| 高丝氨酸激酶 | 苏氨酸 | A20G | [ |
| 苏氨酸脱水酶 | 异亮氨酸 | V323A、H278R‑L351S、F383V、F383A、V140M‑F383A | [ |
| 乙酰羟基酸合成酶 | 异亮氨酸、亮氨酸、缬氨酸 | G20D‑I21D‑I22F | [ |
| 异亮氨酸 | P176S‑D426E‑L57W | [ |
表1 L?异亮氨酸生物合成途径解除反馈抑制位点
Table 1 Feedback inhibition sites of L?isoleucine biosynthesis pathway
| 受底物反馈抑制的酶 | 底物 | 解除反馈抑制位点 | 参考文献 |
|---|---|---|---|
| 天冬氨酸激酶 | 苏氨酸 | N374A、D274A、E278V、G277A、A279V | [ |
| 赖氨酸 | T361A、S381F | [ | |
| 苏氨酸、赖氨酸 | N299L、T336L、M365A、N372A、I272E、Q298G、I375P、E382A、F283R、R384L、S386A、S301F、T308A、F364A、T311I、A279T | [ | |
| 高丝氨酸脱氢酶 | 苏氨酸 | G378E、G378S | [ |
| 高丝氨酸激酶 | 苏氨酸 | A20G | [ |
| 苏氨酸脱水酶 | 异亮氨酸 | V323A、H278R‑L351S、F383V、F383A、V140M‑F383A | [ |
| 乙酰羟基酸合成酶 | 异亮氨酸、亮氨酸、缬氨酸 | G20D‑I21D‑I22F | [ |
| 异亮氨酸 | P176S‑D426E‑L57W | [ |
| 改造策略 | L‑异亮氨酸生产菌株 | L‑异亮氨酸产量/(g·L-1) | 转化率/% | 发酵条件 | 参考文献 |
|---|---|---|---|---|---|
| 过表达关键基因(解除反馈抑制) | K2P55(homG378E‑ilvAF323A) | 14.3 | 13.7 | 分批补料,3 L罐,78 h | [ |
| JHI3‑156/pDXW‑8‑ilvBN1‑ilvA1 | 30.7 | 12.0 | 分批补料,3 L罐,72 h | [ | |
| 修饰转运系统 | YILWΔbrnQ/pXMJ19brnFE | 29.0 | 24.0 | 分批补料,5 L罐,60 h | [ |
| YILWΔalaT | 15.4 | — | 分批补料,3 L罐,72 h | [ | |
| JHI3‑156/pDXW‑8‑lrp‑brnFE | 26.9 | 12.2 | 分批补料,3 L罐,72 h | [ | |
| 辅因子平衡 | IWJ001/pDXW‑8‑gnd‑fbp‑pgl | 29.0 | 13.8 | 分批补料,3 L罐,96 h | [ |
| IWJ001/pDXW‑8‑fusA‑frr‑ilvBN1‑ilvA1‑ppnK | 28.5 | 13.9 | 分批补料,3 L罐,72 h | [ | |
| WM005/pYCW‑1‑ilvBN2‑ppnK1 | 32.1 | 18.1 | 分批补料,5 L罐,72 h | [ |
表2 L?异亮氨酸各改造策略发酵产量
Table 2 Fermentation yield of L?isoleucine by different transformation strategies
| 改造策略 | L‑异亮氨酸生产菌株 | L‑异亮氨酸产量/(g·L-1) | 转化率/% | 发酵条件 | 参考文献 |
|---|---|---|---|---|---|
| 过表达关键基因(解除反馈抑制) | K2P55(homG378E‑ilvAF323A) | 14.3 | 13.7 | 分批补料,3 L罐,78 h | [ |
| JHI3‑156/pDXW‑8‑ilvBN1‑ilvA1 | 30.7 | 12.0 | 分批补料,3 L罐,72 h | [ | |
| 修饰转运系统 | YILWΔbrnQ/pXMJ19brnFE | 29.0 | 24.0 | 分批补料,5 L罐,60 h | [ |
| YILWΔalaT | 15.4 | — | 分批补料,3 L罐,72 h | [ | |
| JHI3‑156/pDXW‑8‑lrp‑brnFE | 26.9 | 12.2 | 分批补料,3 L罐,72 h | [ | |
| 辅因子平衡 | IWJ001/pDXW‑8‑gnd‑fbp‑pgl | 29.0 | 13.8 | 分批补料,3 L罐,96 h | [ |
| IWJ001/pDXW‑8‑fusA‑frr‑ilvBN1‑ilvA1‑ppnK | 28.5 | 13.9 | 分批补料,3 L罐,72 h | [ | |
| WM005/pYCW‑1‑ilvBN2‑ppnK1 | 32.1 | 18.1 | 分批补料,5 L罐,72 h | [ |
| 1 | ELOVARIS R A, HAJISHAFIEE M, ULLRICH S S, et al.. Intragastric administration of leucine and isoleucine does not reduce the glycaemic response to, or slow gastric emptying of, a carbohydrate‑containing drink in type 2 diabetes[J/OL]. Diabetes Res. Clin. Pr., 2021, 171: 108618[2021‑12‑08]. . |
| 2 | ZHAO Y, YAN M Y, JIANG Q, et al.. Isoleucine improved growth performance, and intestinal immunological and physical barrier function of hybrid catfish Pelteobagrus vachelli × Leiocassis longirostris [J]. Fish Shellfish Immun., 2021, 109: 20‑33. |
| 3 | GUILLOUET S, RODAL A A, AN G H, et al.. Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production[J]. Appl. Environ. Microb., 1999, 65(7): 3100‑3107. |
| 4 | D'ESTE M, ALVARADO‑MORALES M, ANGELIDAKI I. Amino acids production focusing on fermentation technologies ‑ a review [J]. Biotechnol. Adv., 2018, 36(1): 14‑25. |
| 5 | LUETKE‑EVERSLOH T, SANTOS C N S, STEPHANOPOULOS G. Perspectives of biotechnological production of L‑tyrosine and its applications[J]. Appl. Microbiol. Biot., 2007, 77(4): 751‑762. |
| 6 | IVANOV K, STOIMENOVA A, OBRESHKOVA D, et al.. Biotechnology in the production of pharmaceutical industry ingredients: amino acids[J]. Biotechnol. Biotec. Eq., 2013, 27(2): 3620‑3626. |
| 7 | EGGELING L, BOTT M. A giant market and a powerful metabolism: L‑lysine provided by Corynebacterium glutamicum [J]. Appl. Microbiol. Biot., 2015, 99(8): 3387‑3394. |
| 8 | YU X L, JIN H Y, LIU W J, et al.. Engineering Corynebacterium glutamicum to produce 5‑aminolevulinic acid from glucose[J/OL]. Microb. Cell Fact., 2015, 14: 183[2021‑12‑08]. . |
| 9 | WANG X. Strategy for improving L‑isoleucine production efficiency in Corynebacterium glutamicum [J]. Appl. Microbiol. Biotechnol., 2019, 103(5): 2101‑2111. |
| 10 | LOTHAR E M. Handbook of Corynebacterium glutamicum [M]. London: Taylor & Francis, 2005. |
| 11 | ZHANG C L, DU S S, LIU Y, et al.. Strategy for enhancing adenosine production under the guidance of transcriptional and metabolite pool analysis[J]. Biotechnol. Lett., 2015, 37(7): 1361‑1369. |
| 12 | BAILEY J E. Toward a science of metabolic engineering[J]. Science, 1991, 252(5013): 1668‑1675. |
| 13 | WANG T, LI Y J, LI J, et al.. An update of the suicide plasmid‑mediated genome editing system in Corynebacterium glutamicum [J]. Microb. Biotechnol., 2019, 12(5): 907‑919. |
| 14 | SUZUKI N, INUI M, YUKAWA H. Site‑directed integration system using a combination of mutant lox sites for Corynebacterium glutamicum [J]. Appl. Microbiol. Biot., 2007, 77(4): 871‑878. |
| 15 | VOGT M, KRUMBACH K, BANG W G, et al.. The contest for precursors: channelling L‑isoleucine synthesis in Corynebacterium glutamicum without byproduct formation[J]. Appl. Microbiol. Biotechnol., 2015, 99(2): 791‑800. |
| 16 | SHIIO I, MIYAJIMA R. Concerted inhibition and its reversal by end products of aspartate kinase in brevibacterium flavum[J]. J. Biochem., 1969, 65(6): 849‑859. |
| 17 | DONG X Y, QUINN P J, WANG X Y. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of L-threonine[J]. Biotechnol. Adv., 2011, 29(1): 11-23. |
| 18 | GUO Y F, XU J Z, HAN M, et al.. Generation of mutant threonine dehydratase and its effects on isoleucine synthesis in Corynebacterium glutamicum [J]. World J. Microb. Biot., 2015, 31(9): 1369‑1377. |
| 19 | CALDER P C. Branched‑chain amino acids and immunity[J]. J. Nutr., 2006, 136(1): 288s‑293s. |
| 20 | GUO Y F, HAN M, YAN W L, et al.. Generation of branched‑chain amino acids resistant Corynebacterium glutamicum acetohydroxy acid synthase by site‑directed mutagenesis[J]. Eiotechnol. Bioproc. E, 2014, 19(3): 456‑467. |
| 21 | CHEN Z, MEYER W Q, RAPPERT S, et al.. Coevolutionary analysis enabled rational deregulation of allosteric enzyme inhibition in Corynebacterium glutamicum for lysine production [J]. Appl. Environ. Microb., 2011, 77(13): 4352‑4360. |
| 22 | YOSHIDA A, TOMITA T, KURIHARA T, et al.. Structural insight into concerted inhibition of alpha2beta2‑type aspartate kinase from Corynebacterium glutamicum [J]. J. Mol. Biol., 2007, 368(2): 521‑36. |
| 23 | SINDELAR G, WENDISCH V F. Improving lysine production by Corynebacterium glutamicum through DNA microarray‑based identification of novel target genes[J]. Appl. Microbiol. Biot., 2007, 76(3): 677‑689. |
| 24 | OHNISHI J, MITSUHASHI S, HAYASHI M, et al.. A novel methodology employing Corynebacterium glutamicum genome information to generate a new L‑lysine‑producing mutant [J]. Appl. Microbiol. Biot., 2002, 58(2): 217‑23. |
| 25 | DONG X Y, ZHAO Y, ZHAO J X, et al.. Characterization of aspartate kinase and homoserine dehydrogenase from Corynebacterium glutamicum IWJ001 and systematic investigation of L‑isoleucine biosynthesis[J]. J. Ind. Microbiol. Biot., 2016, 43(6): 873‑885. |
| 26 | REINSCHEID D J, EIKMANNS B J, SAHM H. Analysis of a Corynebacterium‑glutamicum hom gene coding for a feedback‑resistant homoserine dehydrogenase[J]. J. Bacteriol., 1991, 173(10): 3228‑3230. |
| 27 | PETIT C, KIM Y, LEE S K, et al.. Reduction of feedback inhibition in homoserine kinase (ThrB) of Corynebacterium glutamicum enhances l‑threonine biosynthesis [J]. Acs. Omega., 2018, 3(1): 1178‑1186. |
| 28 | MOCKEL B, EGGELING L, SAHM H. Threonine dehydratases of Corynebacterium‑glutamicum with altered allosteric control ‑ their generation and biochemical and structural‑analysis[J]. Mol. Microbiol., 1994, 13(5): 833‑842. |
| 29 | YIN L, HU X, XU D, et al.. Co‑expression of feedback‑resistant threonine dehydratase and acetohydroxy acid synthase increase L‑isoleucine production in Corynebacterium glutamicum [J]. Metab. Eng., 2012, 14(5): 542‑550. |
| 30 | ELISAKOVA V, PATEK M, HOLATKO J, et al.. Feedback‑ resistant acetohydroxy acid synthase increases valine production in Corynebacterium glutamicum [J]. Appl. Environ. Microb., 2005, 71(1): 207‑213. |
| 31 | ZHANG Y, LIU Y, ZHANG S, et al.. Metabolic engineering of Corynebacterium glutamicum WM001 to improve l‑isoleucine production[J]. Biotechnol. Appl. Biochem., 2020, 68(3): 568‑584. |
| 32 | ZHAO J X, HU X Q, LI Y, et al.. Overexpression of ribosome elongation factor G and recycling factor increases lisoleucine production in Corynebacterium glutamicum [J]. Appl. Microbiol. Biot., 2015, 99(11): 4795‑4805. |
| 33 | WANG J, WEN B, WANG J, et al.. Enhancing L‑isoleucine production by thrABC overexpression combined with alaT deletion in Corynebacterium glutamicum [J]. Appl. Biochem. Biotech., 2013, 171(1): 20‑30. |
| 34 | YIN L, SHI F, HU X, et al.. Increasing L‑isoleucine production in Corynebacterium glutamicum by overexpressing global regulator Lrp and two‑component export system BrnFE [J]. J. Appl. Microbiol., 2013, 114(5): 1369‑1377. |
| 35 | LI N, XU S, DU G C, et al.. Efficient production of L‑homoserine in Corynebacterium glutamicum ATCC 13032 by redistribution of metabolic flux[J/OL]. Biochem. Eng. J., 2020, 161: 107665[2021‑12‑08]. . |
| 36 | EBBIGHAUSEN H, WEIL B, KRAMER R. Transport of branched‑chain amino‑acids in Corynebacterium‑glutamicum [J]. Arch. Microbiol., 1989, 151(3): 238‑244. |
| 37 | TAUCH A, HERMANN T, BURKOVSKI A, et al.. Isoleucine uptake in Corynebacterium glutamicum ATCC 13032 is directed by the brnQ gene product [J]. Arch. Microbiol., 1998, 169(4): 303‑312. |
| 38 | LU J N, BRIGHAM C J, PLASSMEIER J K, et al.. Characterization and modification of enzymes in the 2‑ketoisovalerate biosynthesis pathway of Ralstonia eutropha H16[J]. Appl. Microbiol. Biot., 2015, 99(2): 761‑774. |
| 39 | XIE X, XU L, SHI J, et al.. Effect of transport proteins on L‑isoleucine production with the L‑isoleucine‑producing strain Corynebacterium glutamicum YILW[J]. J. Ind. Microbiol. Biotechnol., 2012, 39(10): 1549‑1556. |
| 40 | BROER S, EGGELING L, KRAMER R. Strains of Corynebacterium‑Glutamicum with different lysine productivities may have different lysine excretion systems[J]. Appl. Environ. Microb., 1993, 59(1): 316‑321. |
| 41 | DONG X Y, ZHAO Y, HU J Y, et al.. Attenuating L‑lysine production by deletion of ddh and lysE and their effect on L‑threonine and L‑isoleucine production in Corynebacterium glutamicum [J]. Enzyme Microb. Tech., 2016, 93‑94:70‑78. |
| 42 | KENNERKNECHT N, SAHM H, YEN M R, et al.. Export of L‑isoleucine from Corynebacterium glutamicum: a two‑gene‑encoded member of a new translocator family[J]. J. Bacteriol., 2002, 184(14): 3947‑3956. |
| 43 | LANGE C, MUSTAFI N, FRUNZKE J, et al.. Lrp of Corynebacterium glutamicum controls expression of the brnFE operon encoding the export system for L‑methionine and branched‑chain amino acids [J]. J. Biotechnol., 2012, 158(4): 231‑241. |
| 44 | TAKENO S, HORI K, OHTANI S, et al.. L‑Lysine production independent of the oxidative pentose phosphate pathway by Corynebacterium glutamicum with the Streptococcus mutans gapN gene[J]. Metab. Eng., 2016, 37: 1‑10. |
| 45 | TAKENO S, MURATA R, KOBAYASHI R, et al.. Engineering of Corynebacterium glutamicum with an NADPH‑generating glycolytic pathway for L‑lysine production[J]. Appl. Environ. Microb., 2010, 76(21): 7154‑7160. |
| 46 | MA W, WANG J, LI Y, et al.. Enhancing pentose phosphate pathway in Corynebacterium glutamicum to improve l‑isoleucine production[J]. Biotechnol. Appl. Biochem., 2016, 63(6): 877‑885. |
| 47 | ZHU S, CAI D, LIU Z, et al.. Enhancement of bacitracin production by NADPH generation via overexpressing glucose‑6‑phosphate dehydrogenase zwf in Bacillus licheniformis [J]. Appl. Biochem. Biotechnol., 2019, 187(4): 1502‑1514. |
| 48 | SHI F, LI K, HUAN X, et al.. Expression of NAD(H) kinase and glucose‑6‑phosphate dehydrogenase improve NADPH supply and L‑isoleucine biosynthesis in Corynebacterium glutamicum ssp. lactofermentum[J]. Appl. Biochem. Biotechnol., 2013, 171(2): 504‑521. |
| 49 | KAWAI S, MORI S, MUKAI T, et al.. Molecular characterization of Escherichia coli NAD kinase[J]. Eur. J. Biochem., 2001, 268(15): 4359‑4365. |
| 50 | SAUER U, CANONACO F, HERI S, et al.. The soluble and membrane‑bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli [J]. J. Biol. Chem., 2004, 279(8): 6613‑6619. |
| 51 | AKINTERINWA O, CIRINO P C. Anaerobic obligatory xylitol production in Escherichia coli strains devoid of native fermentation pathways[J]. Appl. Environ. Microbiol., 2011, 77(2): 706‑709. |
| 52 | RATHNASINGH C, RAJ S M, LEE Y, et al.. Production of 3‑hydroxypropionic acid via malonyl‑CoA pathway using recombinant Escherichia coli strains[J]. J. Biotechnol., 2012, 157(4): 633‑640. |
| 53 | ZHAN M, KAN B, DONG J, et al.. Metabolic engineering of Corynebacterium glutamicum for improved L‑arginine synthesis by enhancing NADPH supply[J]. J. Ind. Microbiol. Biotechnol., 2019, 46(1): 45‑54. |
| 54 | SHI A, ZHU X, LU J, et al.. Activating transhydrogenase and NAD kinase in combination for improving isobutanol production[J]. Metab. Eng., 2013, 16: 1‑10. |
| 55 | WU W, ZHANG Y, LIU D, et al.. Efficient mining of natural NADH‑utilizing dehydrogenases enables systematic cofactor engineering of lysine synthesis pathway of Corynebacterium glutamicum [J]. Metab. Eng., 2019, 52: 77‑86. |
| 56 | XU X, CHEN J, WANG Q, et al.. Mutagenesis of key residues in the binding center of L‑aspartate‑b‑semialdehyde dehydrogenase from Escherichia coli enhances utilization of the cofactor NAD(H)[J]. ChemBioChem, 2016, 17(1): 56‑64. |
| 57 | XU J Z, XIA X H, ZHANG J L, et al.. A method for gene amplification and simultaneous deletion in Corynebacterium glutamicum genome without any genetic markers[J]. Plasmid, 2014, 72: 9‑17. |
| 58 | XU D, TAN Y, HUAN X, et al.. Construction of a novel shuttle vector for use in Brevibacterium flavum, an industrial amino acid producer [J]. J. Microbiol. Methods, 2010, 80(1): 86‑92. |
| 59 | TAUCH A, GOTKER S, PUHLER A, et al.. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains[J]. J. Biotechnol., 2002, 99(1): 79‑91. |
| 60 | IKEDA M, KATSUMATA R. A novel system with positive selection for the chromosomal integration of replicative plasmid DNA in Corynebacterium glutamicum [J]. Microbiology, 1998, 144: 1863‑1868. |
| 61 | LOBET Y, PEACOCK M G, CIEPLAK W. Frame‑shift mutation in the Lacz gene of certain commercially available Puc18 plasmids[J/OL]. Nucl. Acids Res., 1989, 17(12): 4897[2021‑12‑08]. . |
| 62 | XU D, TAN Y, SHI F, et al.. An improved shuttle vector constructed for metabolic engineering research in Corynebacterium glutamicum [J]. Plasmid, 2010, 64(2): 85‑91. |
| 63 | VOGT M, HAAS S, KLAFFL S, et al.. Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for L‑leucine overproduction[J]. Metab. Eng., 2014, 22: 40‑52. |
| 64 | XU J, ZHANG J, HAN M, et al.. A method for simultaneous gene overexpression and inactivation in the Corynebacterium glutamicum genome [J]. J. Ind. Microbiol. Biotechnol., 2016, 43(10): 1417‑1427. |
| 65 | STERNBERG N, SAUER B, HOESS R, et al.. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation[J]. J. Mol. Biol., 1986, 187(2): 197‑212. |
| 66 | GUO F, GOPAUL D N, DUYNE G D. Structure of Cre recombinase complexed with DNA in a site‑specific recombination synapse[J]. Nature, 1997, 389(6646): 40‑46. |
| 67 | HU J, TAN Y, LI Y, et al.. Construction and application of an efficient multiple‑gene‑deletion system in Corynebacterium glutamicum [J]. Plasmid, 2013, 70(3): 303‑313. |
| 68 | SUZUKI N, OKAYAMA S, NONAKA H, et al.. Large‑scale engineering of the Corynebacterium glutamicum genome[J]. Appl. Environ. Microb., 2005, 71(6): 3369‑3372. |
| 69 | HU J, LI Y, ZHANG H, et al.. Construction of a novel expression system for use in Corynebacterium glutamicum [J]. Plasmid, 2014, 75: 18‑26. |
| 70 | 赵传仕, 王忠民, 王德良,等..利用同源重组技术调节啤酒中SO2含量研究[J].酿酒,2006(06):62‑64. |
| 71 | KOS C H. Cre/loxP system for generating tissue‑specific knockout mouse models[J]. Nutr. Rev., 2004, 62(6 Pt 1): 243‑246. |
| 72 | KIM H, KIM M, IM S K, et al.. Mouse Cre‑LoxP system: general principles to determine tissue‑specific roles of target genes[J]. Lab. Anim. Res., 2018, 34(4): 147‑159. |
| 73 | ZHANG Y M, BUCHHOLZ F, MUYRERS J P P, et al.. A new logic for DNA engineering using recombination in Escherichia coli [J]. Nat. Genet., 1998, 20(2): 123‑128. |
| 74 | WANG H H, ISAACS F J, CARR P A, et al.. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894‑898. |
| 75 | MUYRERS J P, ZHANG Y, BENES V, et al.. Point mutation of bacterial artificial chromosomes by ET recombination[J]. EMBO Rep., 2000, 1(3): 239‑243. |
| 76 | NARAYANAN K, WILLIAMSON R, ZHANG Y, et al.. Efficient and precise engineering of a 200 kb beta‑globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system[J]. Gene Ther., 1999, 6(3): 442‑447. |
| 77 | ELLIS H M, YU D, DITIZIO T, et al.. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single‑stranded oligonucleotides [J]. Proc. Natl. Acad. Sci. USA, 2001, 98(12): 6742‑6726. |
| 78 | ZHANG Y, MUYRERS J P, RIENTJES J, et al.. Phage annealing proteins promote oligonucleotide‑directed mutagenesis in Escherichia coli and mouse ES cells[J/OL]. BMC Mol. Biol., 2003, 4(1): 1[2021‑12‑08]. . |
| 79 | HUANG Y Y, LI L, XIE S, et al.. Recombineering using RecET in Corynebacterium glutamicum ATCC14067 via a self‑excisable cassette[J/OL]. BMC Mol. Biol., 2017, 7: 7916[2021‑12‑08]. . |
| 80 | ZHAO N N, LI L, LUO G J, et al.. Multiplex gene editing and large DNA fragment deletion by the CRISPR/Cpf1‑RecE/T system in Corynebacterium glutamicum [J]. J. Ind. Microbiol. Biot., 2020, 47(8): 599‑608. |
| 81 | HEIDER S A E, WENDISCH V F. Engineering microbial cell factories: metabolic engineering of Corynebacterium glutamicum with a focus on non‑natural products [J]. Biotechnol. J., 2015, 10(8): 1170‑1184. |
| 82 | PARK S H, KIM H U, KIM T Y, et al.. Metabolic engineering of Corynebacterium glutamicum for L‑arginine production[J/OL]. Nat. Commun., 2014, 5: 4618[2021‑12‑08]. . |
| 83 | CHO J S, CHOI K R, PRABOWO C P S, et al.. CRISPR/Cas9‑coupled recombineering for metabolic engineering of Corynebacterium glutamicum [J]. Metab. Eng., 2017, 42: 157‑167. |
| 84 | JIANG Y, QIAN F H, YANG J J, et al.. CRISPR‑Cpf1 assisted genome editing of Corynebacterium glutamicum [J/OL]. Nat. Commun., 2017, 8: 15179[2021‑12‑08]. . |
| 85 | SUN B B, YANG J J, YANG S, et al.. A CRISPR‑Cpf1‑assisted non‑homologous end joining genome editing system of Mycobacterium smegmatis [J/OL]. Biotechnol. J., 2018, 13(9): e1700588[2021‑12‑08]. . |
| 86 | 靳海迎.基于CRISPR‑Cas9系统的谷氨酸棒状杆菌基因组编辑技术的研究[D].济南:山东大学,2017. |
| [1] | 张奋强,刘欢,黄丽娜,姜梦嫣,周义杰,邹宇晨,杨明峰,马兰青,. 树莓酮生物合成途径及关键酶功能研究进展[J]. 生物技术进展, 2017, 7(2): 111-115. |
| [2] | 王晓婧,陆伟,徐玉泉,梁晓东. 真菌苯二酚内酯聚酮类化合物生物合成研究进展 [J]. 生物技术进展, 2015, 5(2): 89-94. |
| [3] | 白净,陆伟,徐玉泉,陈明. 苯二酚内酯合成途径中的聚酮合酶组合表达研究[J]. 生物技术进展, 2015, 5(1): 54-59. |
| [4] | 袁志勇,朱红惠,霍光华,冯广达. 微囊藻毒素生物合成及其检测的分子生物学研究进展[J]. 生物技术进展, 2012, 2(5): 328-334. |
| [5] | 宫硖,薛静,张晓东. 植物花青素合成途径中的调控基因研究进展[J]. 生物技术进展, 2011, 1(6): 381-90. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部