


生物技术进展 ›› 2022, Vol. 12 ›› Issue (1): 27-35.DOI: 10.19586/j.2095-2341.2021.0132
王婉洁( ), 陈南珠, 郝海生, 赵学明, 朱化彬, 杜卫华(
), 陈南珠, 郝海生, 赵学明, 朱化彬, 杜卫华( )
)
收稿日期:2021-07-13
接受日期:2021-10-08
出版日期:2022-01-25
发布日期:2022-01-26
通讯作者:
杜卫华
作者简介:王婉洁E-mail:wangwanjie101@163.com;
基金资助:
Wanjie WANG( ), Nanzhu CHEN, Haisheng HAO, Xueming ZHAO, Huabin ZHU, Weihua DU(
), Nanzhu CHEN, Haisheng HAO, Xueming ZHAO, Huabin ZHU, Weihua DU( )
)
Received:2021-07-13
Accepted:2021-10-08
Online:2022-01-25
Published:2022-01-26
Contact:
Weihua DU
摘要:
表观遗传修饰通过改变染色质空间构象和基因表达调控胚胎发育、细胞分化、器官发生和癌症形成,其调控形式包括组蛋白甲基化、组蛋白乙酰化、DNA甲基化、基因印迹和X染色体失活等。缺失的、小的、同源异形2(absent, small, homologous 2,ASH2)是组蛋白赖氨酸甲基转移酶复合物的核心成分,其属于三胸腔结构蛋白家族;在哺乳动物中ASH2可特异性甲基化H3K4,激活基因转录。在介绍组蛋白甲基化和三胸腔结构蛋白的基础上,综述了ASH2甲基酶对基因转录、HOX基因表达、癌症发生发展和细胞分化的调控功能,以期为其在动物繁育和人类疾病治疗中的应用提供思路。
中图分类号:
王婉洁, 陈南珠, 郝海生, 赵学明, 朱化彬, 杜卫华. 组蛋白甲基转移酶ASH2的研究进展[J]. 生物技术进展, 2022, 12(1): 27-35.
Wanjie WANG, Nanzhu CHEN, Haisheng HAO, Xueming ZHAO, Huabin ZHU, Weihua DU. Research Progress of Histone Methyltransferases ASH2[J]. Current Biotechnology, 2022, 12(1): 27-35.
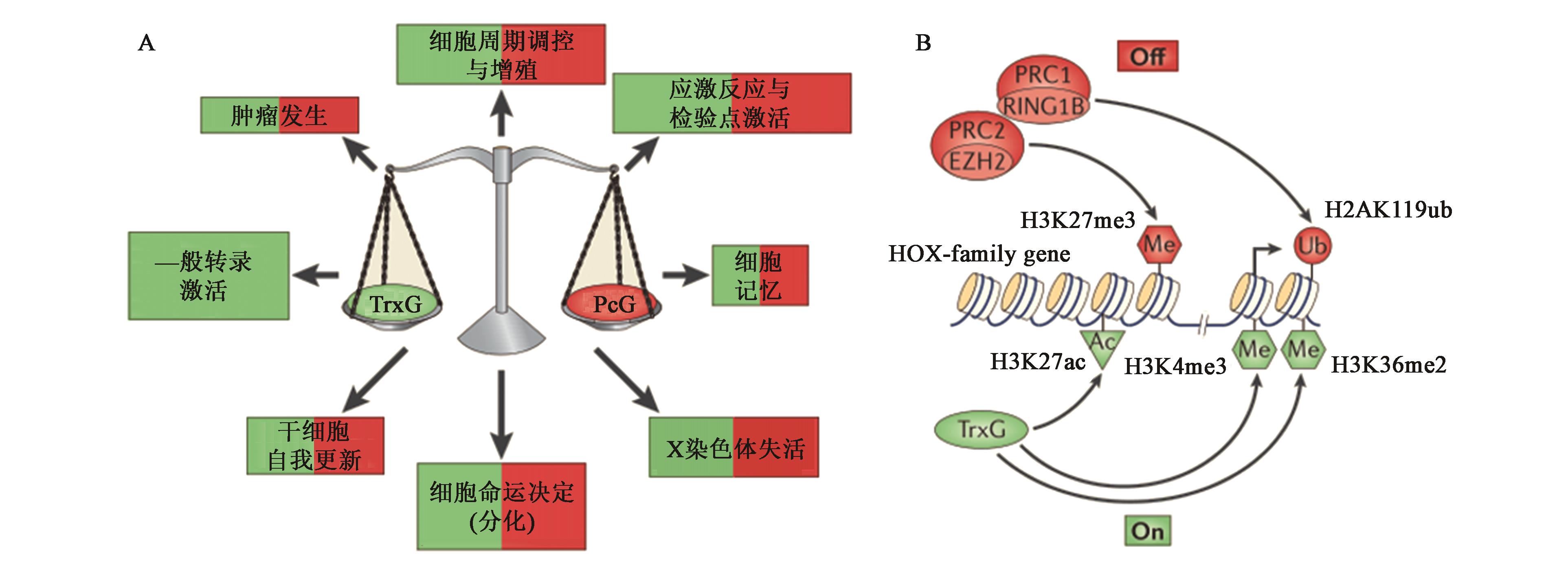
图1 TrxG和PcG相互拮抗的动态平衡[20]A:PcG和TrxG复合物的动态平衡影响多种生物学过程;B:PcG和TrxG复合物通过多种反应元件的调节发挥拮抗作用
Fig.1 Dynamic equilibrium of mutual antagonism between TrxG and PcG[20]
| 亚基 | 复合物 | 哺乳动物同源物 | 分子功能 |
|---|---|---|---|
| PC | PRC1 | CBX2、CBX4、CBX6、CBX7、CBX8 | 通过染色质结构域与H2K27me3结合 |
| PSC | PRC1 | BMI1(PCGF4)、MEL18(PCGF2) | 含锌指结构域蛋白,与DNA和致密染色质结合 |
| PH | PRC1 | PHC1(EDR1)、PHC2(EDR2)、PHC3(EDR3) | 具有锌指结构域,促进蛋白自聚集 |
| SCE(DRING1) | PRC1 | RING1A、RING1B | E3泛素连接酶,单双酯化H2AK118(哺乳动物中的K119) |
| E(Z) | PRC2 | EZH1、EZH2 | 通过SET结构域甲基化H3K27 |
| SU(Z)12 | PRC2 | SUZ12 | 通过VEFS‑box结构域增强E(Z)活性 |
| ESC | PRC2 | EED | 通过WD重复序列促进蛋白间相互作用,通过结合H3K27me3增强抑制作用 |
| P55(NURF55、CAF1) | PRC2 | RBAP46、RBAP48 | 与SU(Z)12和组蛋白相互作用 |
| PHO | PHORC | YY1 | 通过锌指结构特异性地结合DNA序列 |
| SFMBT | PHORC | — | 结合甲基化的H3和H4赖氨酸 |
表1 PcG复合物成分[26]
Table 1 Composition of PcG complex[26]
| 亚基 | 复合物 | 哺乳动物同源物 | 分子功能 |
|---|---|---|---|
| PC | PRC1 | CBX2、CBX4、CBX6、CBX7、CBX8 | 通过染色质结构域与H2K27me3结合 |
| PSC | PRC1 | BMI1(PCGF4)、MEL18(PCGF2) | 含锌指结构域蛋白,与DNA和致密染色质结合 |
| PH | PRC1 | PHC1(EDR1)、PHC2(EDR2)、PHC3(EDR3) | 具有锌指结构域,促进蛋白自聚集 |
| SCE(DRING1) | PRC1 | RING1A、RING1B | E3泛素连接酶,单双酯化H2AK118(哺乳动物中的K119) |
| E(Z) | PRC2 | EZH1、EZH2 | 通过SET结构域甲基化H3K27 |
| SU(Z)12 | PRC2 | SUZ12 | 通过VEFS‑box结构域增强E(Z)活性 |
| ESC | PRC2 | EED | 通过WD重复序列促进蛋白间相互作用,通过结合H3K27me3增强抑制作用 |
| P55(NURF55、CAF1) | PRC2 | RBAP46、RBAP48 | 与SU(Z)12和组蛋白相互作用 |
| PHO | PHORC | YY1 | 通过锌指结构特异性地结合DNA序列 |
| SFMBT | PHORC | — | 结合甲基化的H3和H4赖氨酸 |
| 1 | KOUZARIDES T. Chromatin modifications and their function [J]. Cell, 2007, 128(4): 693-705. |
| 2 | ALLIS C D, JENUWEIN T. The molecular hallmarks of epigenetic control [J]. Nat. Rev. Genet., 2016, 17(8): 487-500. |
| 3 | HYUN K, JEON J, PARK K, et al.. Writing, erasing and reading histone lysine methylations [J/OL]. Exp. Mol. Med., 2017, 49(4): e324[2021-6-29]. . |
| 4 | OSIPOVICH A B, GANGULA R, VIANNA P G, et al.. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation [J]. Development, 2016, 143(24): 4595-4607. |
| 5 | ANDREW J, BANNISTER R S, TONY K. Histone methylation: dynamic or static?[J]. Cell, 2002, 109: 801-806. |
| 6 | CHI P, ALLIS C D, WANG G G. Covalent histone modifications miswritten, misinterpreted and mis-erased in human cancers[J]. Nat. Rev. Cancer, 2010, 10(7): 457-469. |
| 7 | BLACK J C, CVAN RECHEM, WHETSTINE J R. Histone lysine methylation dynamics: establishment, regulation, and biological impact [J]. Mol. Cell, 2012, 48(4): 491-507. |
| 8 | MARCHO C, CUI W, MAGER J. Epigenetic dynamics during preimplantation development[J]. Reproduction, 2015, 150(3): 109-120. |
| 9 | WANG J, ZHOU Y, YIN B, et al.. ASH2L: alternative splicing and downregulation during induced megakaryocytic differentiation of multipotential leukemia cell lines[J]. J. Mol. Med., 2001, 79(7): 399-405. |
| 10 | STOLLER J Z, HUANG L, TAN C C, et al.. Ash2l interacts with Tbx1 and is required during early embryogenesis [J]. Exp. Biol. Med., 2010, 235(5): 569-576. |
| 11 | CHEN Y, CAO F, WAN B B, et al.. Structure of the SPRY domain of human Ash2L and its interactions with RbBP5 and DPY30 [J]. Cell Res., 2012, 22(3): 598-602. |
| 12 | ERFANI P, TOME-GARCIA J, CANOLL P, et al.. EGFR promoter exhibits dynamic histone modifications and binding of ASH2L and P300 in human germinal matrix and gliomas[J]. Epigenetics, 2015, 10(6): 496-507. |
| 13 | ZENG K, WU Y, WANG C Y, et al.. ASH2L is involved in promotion of endometrial cancer progression via upregulation of PAX2 transcription [J]. Cancer Sci., 2020, 111(6): 2062-2077. |
| 14 | BERNSTEIN B E, MIKKELSEN T S, XIE X H, et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells [J]. Cell, 2006, 125(2): 315-326. |
| 15 | LUSCHER-FIRZLAFF J, GAWLISTA I, VERVOORTS J, et al.. The human trithorax protein hASH2 functions as an oncoprotein[J]. Cancer Res., 2008, 68(3): 749-758. |
| 16 | WU Y J, LI L X, LIU L, et al.. ASH2L-promoted HOXC8 gene expression plays a role in mixed lineage leukemia-rearranged acute leukemia [J]. Onco. Targets Ther., 2020, 13: 381-387. |
| 17 | STEFFEN P A, RINGROSE L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory?[J]. Nat. Rev. Mol. Cell Biol., 2014, 15(5): 340-356. |
| 18 | KINGSTON R E, TAMKUN J W. Transcriptional regulation by trithorax-group proteins[J/OL]. Cold Spring Harb. Perspect Biol., 2014, 6(10): a019349[2021-7-10].. |
| 19 | LI L, RUAN X B, WEN C, et al.. The COMPASS family protein ASH2L mediates Corticogenesis via transcriptional regulation of Wnt signaling [J]. Cell Rep., 2019, 28(3): 698-711. |
| 20 | SCHUETTENGRUBER B, MARTINEZ A M, IOVINO N, et al.. Trithorax group proteins: switching genes on and keeping them active [J]. Nat. Rev. Mol. Cell Biol., 2011, 12(12): 799-814. |
| 21 | ANG Y S, TSAI S Y, LEE D F, et al.. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network [J]. Cell, 2011, 145(2): 183-197. |
| 22 | BAGCHI A, PAPAZOGLU C, WU Y, et al.. CHD5 is a tumor suppressor at human 1p36 [J]. Cell, 2007, 128(3): 459-475. |
| 23 | PULLIRSCH D, HARTEL R, KISHIMOTO H, et al.. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation [J]. Development, 2010, 137(6): 935-943. |
| 24 | LIU H, CHENG E H, HSIEH J J. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions [J]. Genes Dev., 2007, 21(19): 2385-2398. |
| 25 | TYAGI S, HERR W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases [J]. Embo. J., 2009, 28(20): 3185-3195. |
| 26 | GEISLER S J, PARO R. Trithorax and Polycomb group-dependent regulation: a tale of opposing activities [J]. Development, 2015, 142(17): 2876-2887. |
| 27 | KASSIS J A, KENNISON J A, TAMKUN J W. Polycomb and Trithorax group genes in Drosophila [J]. Genetics, 2017, 206(4): 1699-1725. |
| 28 | DI CROCE L, HELIN K. Transcriptional regulation by polycomb group proteins [J]. Nat. Struct. Mol. Biol., 2013, 20(10): 1147-1155. |
| 29 | GROSSNIKLAUS U, PARO R. Transcriptional silencing by polycomb-group proteins [J/OL]. Cold Spring Harb. Perspect Biol., 2014, 6(11): a019331[2021-7-2].. |
| 30 | KU M, KOCHE R P, RHEINBAY E, et al.. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains [J/OL]. PLoS Genet., 2008, 4(10): e1000242[2021-6-15].. |
| 31 | SCHNEIDER R, BANNISTER A J, MYERS F A, et al.. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes[J]. Nat. Cell Biol., 2004, 6(1): 73-77. |
| 32 | SCHNEIDER J, WOOD A, LEE J S, et al.. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression [J]. Mol. Cell, 2005, 19(6): 849-856. |
| 33 | KRIVTSOV A V, ARMSTRONG S A. MLL translocations, histone modifications and leukaemia stem-cell development[J]. Nat. Rev. Cancer, 2007, 7(11): 823-833. |
| 34 | NULAND R, SMITS A H, PALLAKI P, et al.. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes [J]. Mol.Cell Biol., 2013, 33(10): 2067-2077. |
| 35 | KROGAN N J, DOVER J, KHORRAMI S, et al.. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression [J]. J. Biol. Chem., 2002, 277(13): 10753-10755. |
| 36 | BROWN C A, MURRAY A W, VERSTREPEN K J. Rapid expansion and functional divergence of subtelomeric gene families in yeasts [J]. Curr. Biol., 2010, 20(10): 895-903. |
| 37 | LEE J S, SHUKLA A, SCHNEIDER J, et al.. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS [J]. Cell, 2007, 131(6): 1084-1096. |
| 38 | LUKITO Y, CHUJO T, HALE T K, et al.. Regulation of subtelomeric fungal secondary metabolite genes by H3K4me3 regulators CclA and KdmB [J]. Mol. Microbiol., 2019, 112(3): 837-853. |
| 39 | NISLOW C, RAY E, PILLUS L. SET1, A yeast member of the Trithorax family, functions in transcriptional silencing and diverse cellular processes[J]. Mol. Biol. Cell, 1997, 8: 2421-2436. |
| 40 | POKHOLOK D K, HARBISON C T, LEVINE S, et al.. Genome-wide map of nucleosome acetylation and methylation in yeast[J]. Cell, 2005, 122(4): 517-527. |
| 41 | DOUILLET D, SZE C C, RYAN C, et al.. Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, polycomb and DNA methylation [J]. Nat. Genet., 2020, 52(6): 615-625. |
| 42 | RUGG-GUNN P J, COX B J, RALSTON A, et al.. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo [J]. Proc. Natl. Acad. Sci. USA, 2010, 107(24): 10783-10790. |
| 43 | BOCHYNSKA A, LUSCHER-FIRZLAFF J, LUSCHER B. Modes of interaction of KMT2 histone H3 lysine 4 methyltransferase/COMPASS complexes with chromatin [J/OL]. Cell, 2018, 7(3): 17[2021-7-14].. |
| 44 | VERMEULEN M T H. Grasping trimethylation of histone H3 at lysine 4[J]. Epigenomics, 2010, 2(3): 395-406. |
| 45 | MILNE T A, BRIGGS S D, BROCK H W, et al.. MLL targets SET domain methyltransferase activity to Hox gene promoters [J]. Mol. Cell, 2002, 10(5): 1107-1117. |
| 46 | XUE H, YAO T, CAO M, et al.. Structural basis of nucleosome recognition and modification by MLL methyltransferases[J]. Nature, 2019, 573(7774): 445-449. |
| 47 | WAN M, LIANG J, XIONG Y, et al.. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells [J]. J. Biol. Chem., 2013, 288(7): 5039-5048. |
| 48 | DILLON S C, ZHANG X, TRIEVEL R C, et al.. The SET-domain protein superfamily: protein lysine methyltransferases[J/OL]. Genome Biol., 2005, 6(8): 227[2021-8-20].. |
| 49 | DORIGHI K M, SWIGUT T, HENRIQUES T, et al.. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation[J]. Mol. Cell, 2017, 66(4): 568-576. |
| 50 | STEWARD M M, LEE J S, O'DONOVAN A, et al.. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes [J]. Nat. Struct. Mol. Biol., 2006, 13(9): 852-854. |
| 51 | BELTRAN S, BLANCO E, SERRAS F, et al.. Transcriptional network controlled by the trithorax-group gene ash2 in Drosophila melanogaster [J]. Proc. Natl. Acad. Sci. USA, 2003, 100(6): 3293-3298. |
| 52 | GREER E L, MAURES T J, UCAR D, et al.. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans[J]. Nature, 2011, 479(7373): 365-371. |
| 53 | CHEN Y, WAN B, WANG K C, et al.. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding [J]. EMBO Rep., 2011, 12(8): 797-803. |
| 54 | RAMPALLI S, LI L, MAK E, et al.. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation [J]. Nat. Struct. Mol. Biol., 2007, 14(12): 1150-1156. |
| 55 | MALLO M, WELLIK D M, DESCHAMPS J. Hox genes and regional patterning of the vertebrate body plan[J]. Dev. Biol., 2010, 344(1): 7-15. |
| 56 | 王吉伟.HOX基因转录调节机制的研究进展[J].国外医学,2000,20(1):44-46. |
| 57 | TAKAHASHI K, YAMANAK S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors [J]. Cell, 2006, 126(4): 663-676. |
| 58 | 吴雨洁.ASH2L在MLL基因重排急性白血病对HOXC8基因的组蛋白修饰调控机制研究[D].南京:南京医科大学,2016. |
| 59 | XIAO Y, BEDET C, ROBERT V J, et al.. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells [J]. Proc. Natl. Acad. Sci. USA, 2011, 108(20): 8305-8310. |
| 60 | LI Y, GUO Z, CHEN H, et al.. HOXC8-dependent Cadherin 11 expression facilitates breast cancer cell migration through Trio and Rac[J]. Genes Cancer, 2011, 2(9): 880-888. |
| 61 | TAN C C, SINDHU K V, LI S, et al.. Transcription factor Ap2delta associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription [J]. Proc. Natl. Acad. Sci. USA, 2008, 105(21):7472-7477. |
| 62 | ARMSTRONG S A, STAUNTON J E, SILVERMAN L B, et al.. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia [J]. Nat. Genet., 2002, 30(1): 41-47. |
| 63 | NAYAK A, VIALE-BOURONCLE S, MORSCZECK C, et al.. The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation[J]. Mol. Cell, 2014, 55(1): 47-58. |
| 64 | SUN B, TU J, LIANG Q, et al.. Expression of mammalian ASH1 and ASH4 in Drosophila reveals opposing functional roles in neurogenesis [J]. Gene, 2019, 688: 132-139. |
| 65 | ROCHA-VIEGAS L, VILLA R, GUTIERREZ A, et al.. Role of UTX in retinoic acid receptor-mediated gene regulation in leukemia [J]. Mol. Cell Biol., 2014, 34(19): 3765-3775. |
| 66 | ROCHA-VIEGAS L, SILBERMINS M, OGARA M F, et al.. Glucocorticoids uncover a critical role for ASH2L on BCL-X expression regulation in leukemia cells[J/OL]. Biochim. Biophys. Acta. Gene Regul. Mech., 2020, 1863(1): 194475[2021-6-24].. |
| 67 | QI J, HUO L, ZHU Y T, et al.. Absent, small or homeotic 2-like protein (ASH2L) enhances the transcription of the estrogen receptor alpha gene through GATA-binding protein 3 (GATA3) [J]. J. Biol. Chem., 2014, 289(45): 31373-31381. |
| 68 | 孙超,孙士莹,詹磊,等.ASH2L在上皮性卵巢癌中的表达及其对SKOV-3细胞系增殖的影响[J].安徽医科大学学报,2021,56(3):445-448. |
| 69 | MUNGAMURI S K, WANG S, MANFREDI J J, et al.. Ash2L enables P53-dependent apoptosis by favoring stable transcription pre-initiation complex formation on its pro-apoptotic target promoters [J]. Oncogene, 2015, 34(19): 2461-2470. |
| 70 | BURTON A, TORRES-PADILLA M E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis [J]. Nat. Rev. Mol. Cell Biol., 2014, 15(11): 723-734. |
| 71 | ZHOU Y, ZHENG L, LI F, et al.. Bivalent histone codes on WNT5A during odontogenic differentiation [J]. J. Dent. Res., 2018, 97(1): 99-107. |
| [1] | 苗瑞菊, 丁尊丹, 田健, 张红兵, 关菲菲. PET水解酶传统与智能分子设计研究进展[J]. 生物技术进展, 2023, 13(1): 46-54. |
| [2] | 宋怡菲, 谢飞, 马晨, 马雪梅. 高等植物氢化酶活性研究进展[J]. 生物技术进展, 2022, 12(4): 481-489. |
| [3] | 吕海超, 贾哲康, 张文, 蒋丽雯, 晁玉文, 窦文芳. 尿苷二磷酸糖固定化酶合成法研究[J]. 生物技术进展, 2022, 12(2): 270-280. |
| [4] | 张文静, 佟晔, 杨锡文, 曹彦金, 魏计东. 高产中性蛋白酶菌株的筛选、优化及中试放大[J]. 生物技术进展, 2022, 12(1): 112-119. |
| [5] | 张亚格, 庞雨, 张维, 周正富. 脂肪酶研发态势的全球专利分析[J]. 生物技术进展, 2021, 11(6): 749-757. |
| [6] | 孙卉, 张春义, 姜凌. 辅酶Ⅰ体内代谢调控研究进展[J]. 生物技术进展, 2021, 11(4): 526-534. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部