


生物技术进展 ›› 2024, Vol. 14 ›› Issue (3): 422-432.DOI: 10.19586/j.2095-2341.2024.0013
• 研究论文 • 上一篇
收稿日期:2024-01-23
接受日期:2024-02-27
出版日期:2024-05-25
发布日期:2024-06-18
通讯作者:
郭维
作者简介:席凯飞 E-mail: kaifeix@163.com;
基金资助:
Kaifei XI( ), Chengjie LI, Yi DING, Wei GUO(
), Chengjie LI, Yi DING, Wei GUO( )
)
Received:2024-01-23
Accepted:2024-02-27
Online:2024-05-25
Published:2024-06-18
Contact:
Wei GUO
摘要:
拟轮枝镰孢菌(Fusarium verticillioides)是引起玉米茎基腐病和穗粒腐病的主要病原菌之一,严重威胁玉米的产量和品质。为了深入研究拟轮枝镰孢菌致病基因的功能,对该菌中非同源末端连接(non-homologous end joining,NHEJ)途径中的2个关键基因FvKu70和FvKu80分别进行了基因敲除以创制高效的基因敲除菌株,并比较了野生型菌株和突变体菌株在营养生长速率、菌落形态、产孢量、对玉米的致病力和基因敲除效率等方面的差异。研究结果表明,FvKu70和FvKu80的基因缺失突变体与野生型FvLNF15-11相比,在PDA平板上的形态特征(如菌丝形态、生长速率、菌落直径、产孢量)没有明显差异,对玉米茎秆的致病力也类似。此外,选择尿嘧啶生物合成相关基因FvpyrG作为敲除的靶基因,分析了FvKu70或FvKu80缺失突变体菌株的同源重组效率,结果显示突变体菌株均显著高于野生型,其中ΔFvKu70的同源重组效率最高。综上所述,FvKu70或FvKu80基因缺失突变体可以快速又高效地实现拟轮枝镰孢菌的基因敲除,为进一步研究该菌的功能基因提供了技术支持。
中图分类号:
席凯飞, 李成杰, 丁艺, 郭维. 利用非同源末端连接缺陷构建拟轮枝镰孢菌的高效基因敲除方法[J]. 生物技术进展, 2024, 14(3): 422-432.
Kaifei XI, Chengjie LI, Yi DING, Wei GUO. Highly Efficient Gene Knockout Method in Fusarium verticillioides Using Nonhomologous End-joining Deficiency[J]. Current Biotechnology, 2024, 14(3): 422-432.
| 引物 | 引物序列(5’→3’) |
|---|---|
| 1F | 5’-GATTACGAATTCGAGCTCGGTACCACTCCTTGTTGGTCTTGCCT-3’ |
| 1R | 5’-ATCTCTAGAGGATCCCCGGGTACCATTGATACGATTGATGTCTG-3’ |
| 2F | 5’-GTCGACCTGCAGGCATGCAAGCTTGACTGAGGTCGAAGGGCAAA-3’ |
| 2R | 5’-AAAACGACGGCCAGTGCCAAGCTTAATATGTAAAAGTCCAACCC-3’ |
| 3F | 5’-GATTACGAATTCGAGCTCGGTACCCTCCGATACCAAGCCAGCA-3’ |
| 3R | 5’-ATCTCTAGAGGATCCCCGGGTACCTCATACCCATCATCGTCTTG-3’ |
| 4F | 5’-GTCGACCTGCAGGCATGCAAGCTTGAGTGAATCATAGGGCCTTG-3’ |
| 4R | 5’-AAAACGACGGCCAGTGCCAAGCTTACATCTCTGGGTGTTGCGAA-3’ |
| 7F | 5’-GGGTAAGGATCACCTTGATA-3’ |
| 7R | 5’-TTAAAGGCAATCGCGATCGA-3’ |
| 8F | 5’-TCCATCAGCATACCACTCCT-3’ |
| 8R | 5’-ATAGACTCGAAGATGGTGAG-3’ |
| HYR | 5’-GTATTGACCGATTCCTTGCGGTCCGAA-3’ |
| YGF | 5’-GATGTAGGAGGGCGTGGATATGTCCT-3’ |
| HYF | 5’-ATGAAAAAGCCTGAACTC-3’ |
| YGR | 5’-TTCCTTTGCCCTCGGACG-3’ |
| Hyg-F | 5’-ATGAAAAAGCCTGAACTCAC-3’ |
| Hyg-R | 5’-CTATTCCTTTGCCCTCGGAC-3’ |
| Ku70F | 5’-GATGACTTGGGTGACATTTC-3’ |
| Ku70R | 5’-GTCCAAAGGAGGAAACTGGT-3’ |
| Ku80F | 5’-GGAGACGAGAAATTCGACCT-3’ |
| Ku80R | 5’-CTGAAGAATTCTGTAGTGCC-3’ |
| pyrG-up-F | 5’-GATTACGAATTCGAGCTCGGTACCCATAAAAACCATGAACCATT-3’ |
| pyrG-up-R | 5’-ATCTCTAGAGGATCCCCGGGTACCGAGGTGAGAGGTGAGAGGTG-3’ |
| pyrG-dw-F | 5’-GTCGACCTGCAGGCATGCAAGCTTTGGAAGTCGGGACGGCCTTG-3’ |
| pyrG-dw-R | 5’-AAAACGACGGCCAGTGCCAAGCTTAGCCTTGTACCTTTCCTTGA-3’ |
| pyrG-u1300-F | 5’-TAACCGTCTCAGACCTGAAAG-3’ |
| pyrG-d1300-R | 5’-TCGGATAGTACTGCCAGTTGA-3’ |
| pyrG-in-F | 5’-CAAGGCCTCGGTTGCATCTC-3’ |
| pyrG-in-R | 5’-CCATGTCATAATGCGTCTTGA-3’ |
| pyrG-F/ | 5’-CGACATTCCACCCATTTACGC-3’ |
| pyrG-R | 5’-GACGTGGTGAATCGGCCGACT-3’ |
表1 本研究所用的引物
Table 1 Primers used in this study
| 引物 | 引物序列(5’→3’) |
|---|---|
| 1F | 5’-GATTACGAATTCGAGCTCGGTACCACTCCTTGTTGGTCTTGCCT-3’ |
| 1R | 5’-ATCTCTAGAGGATCCCCGGGTACCATTGATACGATTGATGTCTG-3’ |
| 2F | 5’-GTCGACCTGCAGGCATGCAAGCTTGACTGAGGTCGAAGGGCAAA-3’ |
| 2R | 5’-AAAACGACGGCCAGTGCCAAGCTTAATATGTAAAAGTCCAACCC-3’ |
| 3F | 5’-GATTACGAATTCGAGCTCGGTACCCTCCGATACCAAGCCAGCA-3’ |
| 3R | 5’-ATCTCTAGAGGATCCCCGGGTACCTCATACCCATCATCGTCTTG-3’ |
| 4F | 5’-GTCGACCTGCAGGCATGCAAGCTTGAGTGAATCATAGGGCCTTG-3’ |
| 4R | 5’-AAAACGACGGCCAGTGCCAAGCTTACATCTCTGGGTGTTGCGAA-3’ |
| 7F | 5’-GGGTAAGGATCACCTTGATA-3’ |
| 7R | 5’-TTAAAGGCAATCGCGATCGA-3’ |
| 8F | 5’-TCCATCAGCATACCACTCCT-3’ |
| 8R | 5’-ATAGACTCGAAGATGGTGAG-3’ |
| HYR | 5’-GTATTGACCGATTCCTTGCGGTCCGAA-3’ |
| YGF | 5’-GATGTAGGAGGGCGTGGATATGTCCT-3’ |
| HYF | 5’-ATGAAAAAGCCTGAACTC-3’ |
| YGR | 5’-TTCCTTTGCCCTCGGACG-3’ |
| Hyg-F | 5’-ATGAAAAAGCCTGAACTCAC-3’ |
| Hyg-R | 5’-CTATTCCTTTGCCCTCGGAC-3’ |
| Ku70F | 5’-GATGACTTGGGTGACATTTC-3’ |
| Ku70R | 5’-GTCCAAAGGAGGAAACTGGT-3’ |
| Ku80F | 5’-GGAGACGAGAAATTCGACCT-3’ |
| Ku80R | 5’-CTGAAGAATTCTGTAGTGCC-3’ |
| pyrG-up-F | 5’-GATTACGAATTCGAGCTCGGTACCCATAAAAACCATGAACCATT-3’ |
| pyrG-up-R | 5’-ATCTCTAGAGGATCCCCGGGTACCGAGGTGAGAGGTGAGAGGTG-3’ |
| pyrG-dw-F | 5’-GTCGACCTGCAGGCATGCAAGCTTTGGAAGTCGGGACGGCCTTG-3’ |
| pyrG-dw-R | 5’-AAAACGACGGCCAGTGCCAAGCTTAGCCTTGTACCTTTCCTTGA-3’ |
| pyrG-u1300-F | 5’-TAACCGTCTCAGACCTGAAAG-3’ |
| pyrG-d1300-R | 5’-TCGGATAGTACTGCCAGTTGA-3’ |
| pyrG-in-F | 5’-CAAGGCCTCGGTTGCATCTC-3’ |
| pyrG-in-R | 5’-CCATGTCATAATGCGTCTTGA-3’ |
| pyrG-F/ | 5’-CGACATTCCACCCATTTACGC-3’ |
| pyrG-R | 5’-GACGTGGTGAATCGGCCGACT-3’ |

图1 FvKu70及其同源蛋白进化树分析注:Fusarium verticilllioides—拟轮枝镰孢菌; Neurospora crassa—粗糙脉孢霉; N. crassa—粗糙脉孢霉; Chaetomium globosum—球毛壳菌; Pyricularia grisea—稻瘟病; Phaeoacremonium minimum—褐枝顶孢霉; Verticillium dahlia—大丽轮枝菌; Lecanicillium saksenae—刀孢蜡蚧菌; Botrytis cinerea—灰霉菌; Sclerotinia sclerotiorum—核盘菌; Macrophomina phaseolina—菜豆壳球孢菌; Parastagonospora nodorum—旁星孢杆菌; Coccidioides immitis—粗球孢子菌; Blastomyces gilchristii—吉氏芽生菌; Aspergillus clavatus—棒曲霉; A. fumigatus—烟曲霉; A. terreus—土曲霉; A. oryzae—米曲霉; A. sojae—酱油曲霉; A. parasiticus—寄生曲霉; Saccharomyces cerevisiae—酿酒酵母。
Fig. 1 The phylogenetic tree of FvKu70 and its homologous proteins

图2 FvKu80及其同源蛋白进化树分析注:Fusarium verticilllioides—拟轮枝镰孢菌; Lecanicillium saksenae—刀孢蜡蚧菌; Verticillium dahliae—大丽轮枝菌; Phaeoacremonium minimum—褐枝顶孢霉; Chaetomium globosum—球毛壳菌; Neurospora crassa—粗糙脉孢霉; N. crassa—粗糙脉孢霉; Pyricularia grisea—稻瘟菌; Botrytis cinerea—灰霉菌; Sclerotinia sclerotiorum—核盘菌; Parastagonospora nodorum—旁星孢杆菌; Macrophomina phaseolina—菜豆壳球孢菌; Coccidioides immitis—粗球孢子菌; Blastomyces gilchristii—吉氏芽生菌; Aspergillus clavatus—棒曲霉; A. fumigatus—烟曲霉; A. terreus—土曲霉; A. oryzae—米曲霉; A. sojae—酱油曲霉; A. parasiticus—寄生曲霉; Saccharomyces cereuisiae—酿酒酵母。
Fig. 2 The phylogenetic tree was constructed to analyze the evolutionary relationships between FvKu80 and its homologous proteins.
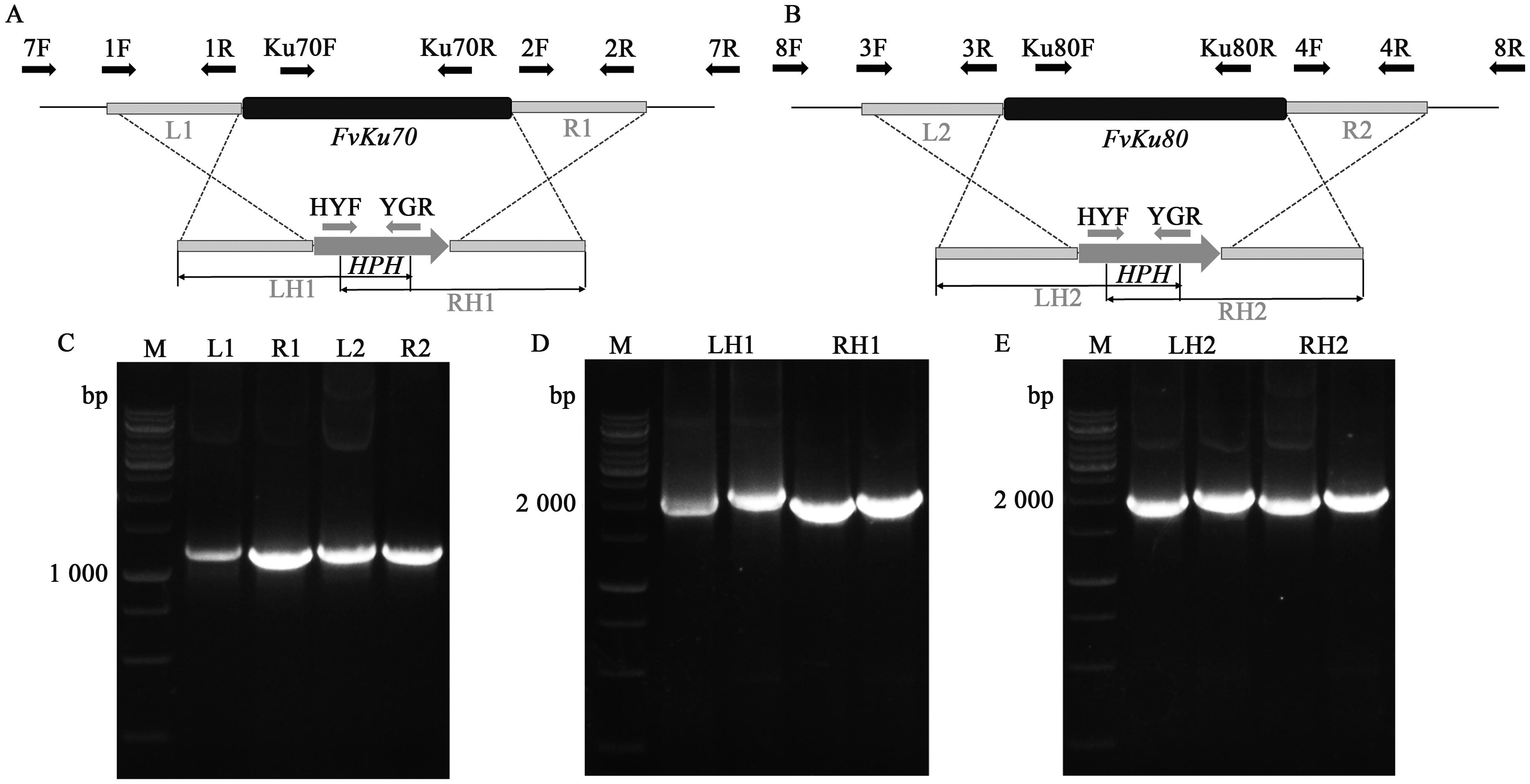
图3 FvKu70和FvKu80基因敲除示意图和载体构建A:FvKu70基因敲除示意图;B:FvKu80基因敲除示意图;C:FvKu70的上下游片段L1/R1和FvKu80基因的上下游片段L2/R2;D:扩增用于Split敲除FvKu70时导入原生质体的片段LH1和RH1;E:扩增用于Split敲除FvKu80时导入原生质体的片段LH2和RH2;M—DNA maker
Fig. 3 The gene knockout diagram and construction of knock-out cassette of FvKu70 and FvKu80
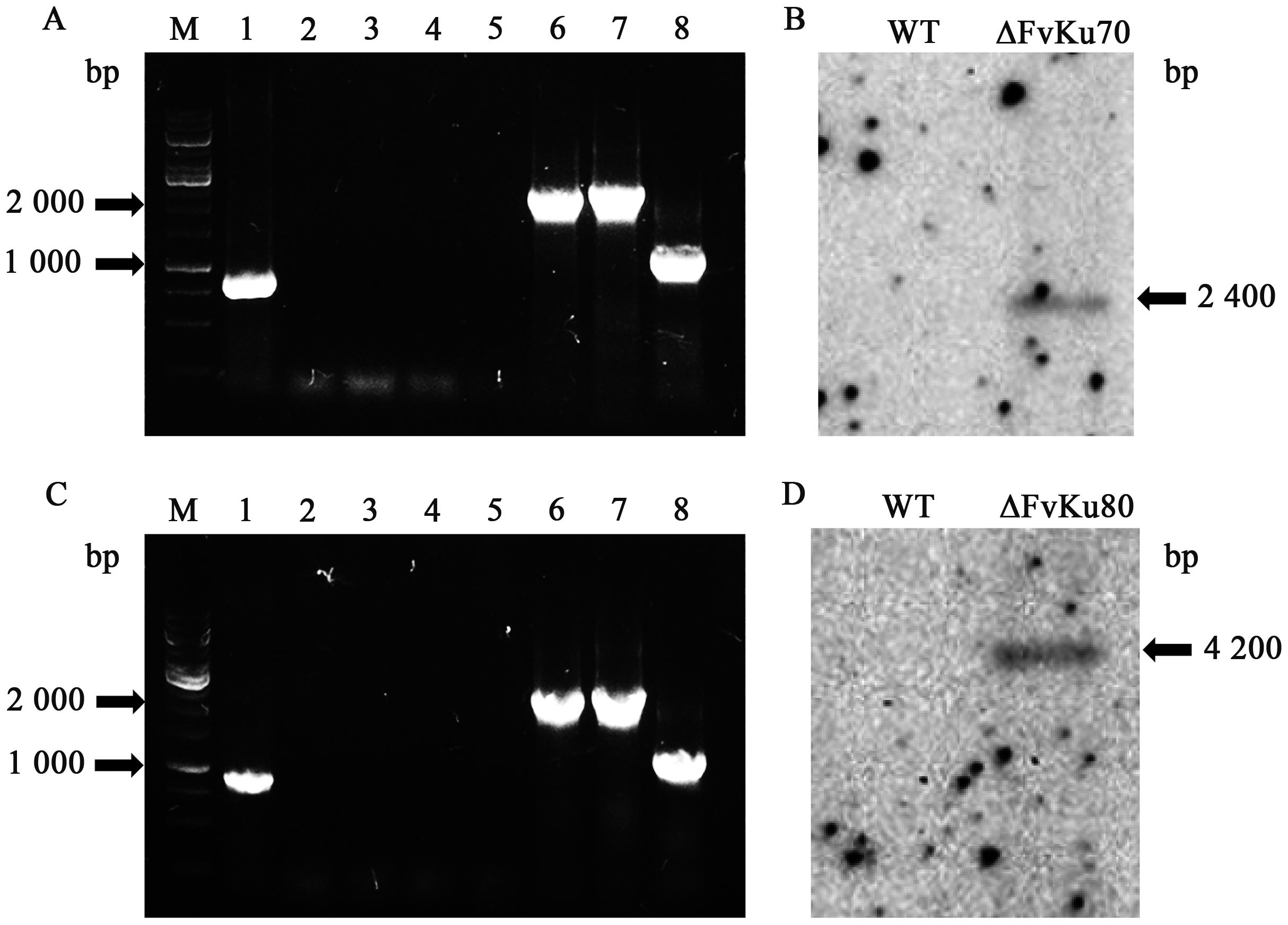
图4 ΔFvKu70和ΔFvKu80的PCR及Southern blot鉴定A: ΔFvKu70的PCR检测,1~4分别为利用Ku70F/R、7F/YGR、HYF/7R和HYF/YGR检测野生型菌株基因组中的目标条带,5~8分别为利用Ku70F/R、7F/YGR、HYF/7R和HYF/YGR检测突变体菌株基因组中的目标条带;B:ΔFvKu70的Southern blot验证结果;C:ΔFvKu80的PCR检测,1~4分别为利用Ku80F/R、8F/YGR、HYF/8R和HYF/YGR检测野生型菌株基因组中的目标条带,5~8分别为利用Ku80F/R、8F/YGR、HYF/8R和HYF/YGR检测突变体菌株基因组中的目标条带;D:ΔFvKu80的Southern blot验证结果;M—DNA maker
Fig. 4 PCR and Southern blot identification of ΔFvKu70 and ΔFvKu80
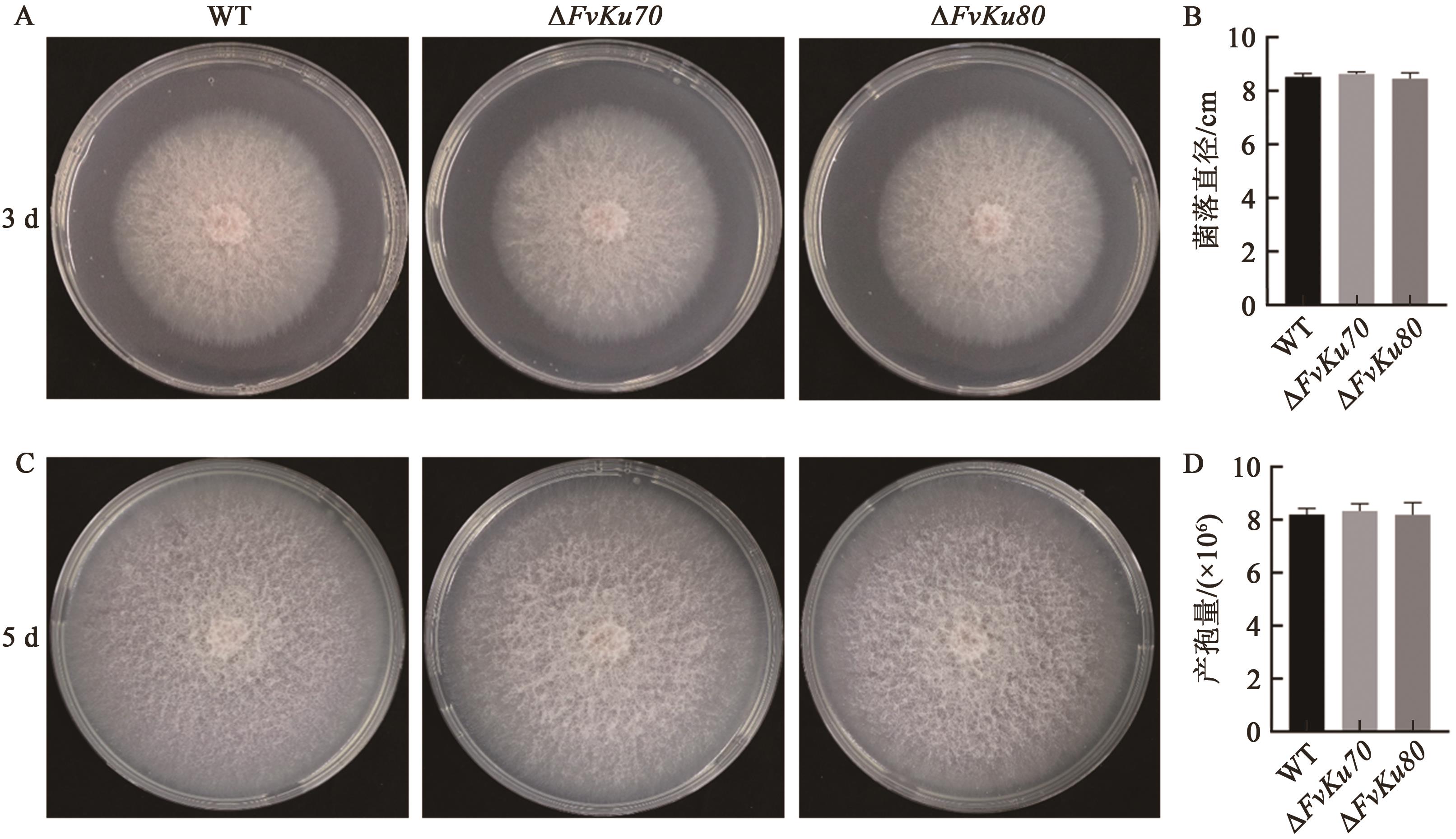
图5 ΔFvKu70和ΔFvKu80基因敲除菌株生长速率测定及菌落形态A: FvLNF15-11、ΔFvKu70和ΔFvKu80在第3天的菌落形态图;B:FvLNF15-11、ΔFvKu70和ΔFvKu80在第5天的菌落直径测量结果;C: FvLNF15-11、ΔFvKu70和ΔFvKu80在第5天的菌落形态图;D:FvLNF15-11、ΔFvKu70和ΔFvKu80培养5 d后的产孢量测定结果
Fig. 5 Assessment of vegetative growth rate and colony morphology in ΔFvKuU70 and ΔFvKu80

图6 ΔFvKu70和ΔFvKu80的致病力测定A: 玉米茎秆分别接种FvLNF15-11、ΔFvKu70和ΔFvKu80 7 d后的症状;B:玉米茎秆分别接种FvLNF15-11、ΔFvKu70和ΔFvKu80 7 d后的病斑面积
Fig. 6 Pathogenicity analysis of ΔFvKu70 and ΔFvKu80 on maize stalks
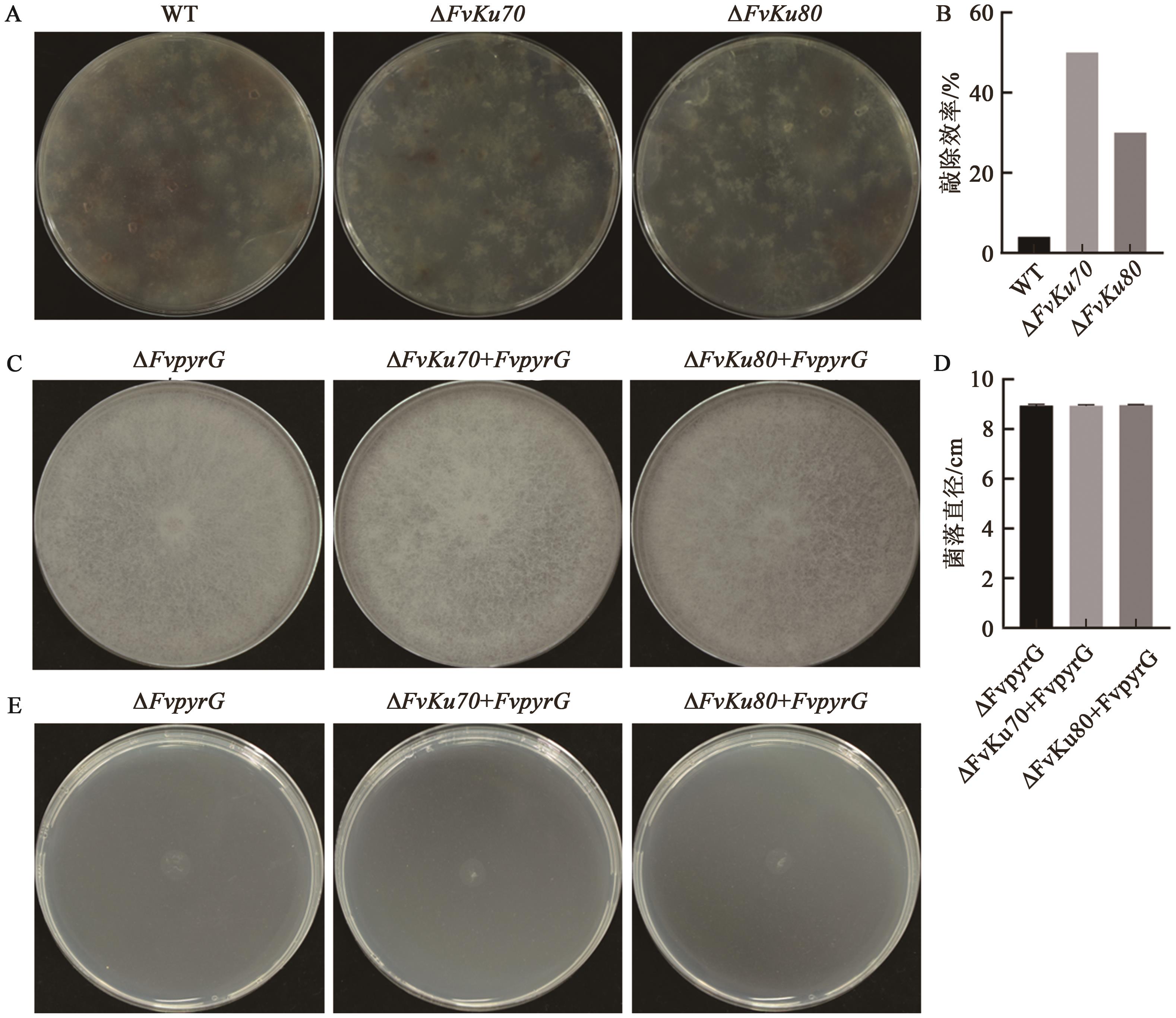
图7 分别以FvLNF15-11、ΔFvKu70和ΔFvKu80为受体菌敲除FvpyrG的效率评估A: 以FvLNF15-11、ΔFvKu70和ΔFvKu80为受体菌敲除FvpyrG基因的转化平板;B:以FvLNF15-11、ΔFvKu70和ΔFvKu80为受体菌敲除FvpyrG基因的敲除效率;C:以FvLNF15-11、ΔFvKu70和ΔFvKu80为受体菌敲除FvpyrG基因后菌落在添加尿嘧啶和尿嘧啶核苷的MM平板上的生长表型;D:对图C的菌落生长直径测量结果;E:以FvLNF15-11、ΔFvKu70和ΔFvKu80为受体菌敲除FvpyrG基因后菌落在不添加尿嘧啶和尿嘧啶核苷的MM平板上的生长情况
Fig. 7 Evaluation of FvpyrG gene knockout efficiency using FvLNF15-11, ΔFvKu70, and ΔFvKu80 as recipient strains
| 1 | KAMLE M, MAHATO D K, DEVI S, et al.. Fumonisins: impact on agriculture, food, and human health and their management strategies[J/OL]. Toxins, 2019, 11(6): 328[2023-10-20]. . |
| 2 | JONES R E, HUMPHREY T C. Homologous recombination and nonhomologous end-joining repair in yeast[M]. 2nd ed. New York: Academic Press, 2019: 2715-2726. |
| 3 | HABER J E. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast[J]. DNA Repair, 2006, 5(9-10): 998-1009. |
| 4 | KITO H, FUJIKAWA T, MORIWAKI A, et al.. MgLig4, a homolog of Neurospora crassa Mus-53 (DNA ligase IV), is involved in, but not essential for, non-homologous end-joining events in Magnaporthe grisea [J]. Fungal Genet. Biol., 2008, 45(12): 1543-1551. |
| 5 | XU D, ZHAO H. Pathway choice for DNA double strand break repair[J]. Sci. Sin. Vitae, 2021, 51(1): 56-69. |
| 6 | GUHA S, BHAUMIK S R. Transcription-coupled DNA double-strand break repair[J/OL]. DNA Repair, 2022, 109: 103211[2024-04-08]. . |
| 7 | D'ADDA DI FAGAGNA F, HANDE M P, TONG W M, et al.. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells[J]. Curr. Biol., 2001, 11(15): 1192-1196. |
| 8 | PRYOR J M, CONLIN M P, CARVAJAL-GARCIA J, et al.. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining[J]. Science, 2018, 361(6407): 1126-1129. |
| 9 | NINOMIYA Y, SUZUKI K, ISHII C, et al.. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining[J]. Proc. Natl. Acad. Sci. USA, 2004, 101(33): 12248-12253. |
| 10 | WEYDA I, YANG L, VANG J, et al.. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR/Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius [J]. J. Microbiol. Meth., 2017, 135: 26-34. |
| 11 | KRAPPMANN S, SASSE C, BRAUS G H. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background[J]. Eukaryot. Cell, 2006, 5(1): 212-215. |
| 12 | MEYER V, ARENTSHORST M, EL-GHEZAL A, et al.. Highly efficient gene targeting in the Aspergillus niger kusA mutant[J]. J. Biotechnol., 2007, 128(4): 770-775. |
| 13 | VILLALBA F, COLLEMARE J, LANDRAUD P, et al.. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining[J]. Fungal Genet. Biol., 2008, 45(1): 68-75. |
| 14 | CHANG P K, SCHARFENSTEIN L L, WEI Q, et al.. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus [J]. J. Microbiol. Meth., 2010, 81(3): 240-246. |
| 15 | PÖGGELER S, KÜCK U. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian Ku70 ortholog[J]. Gene, 2006, 378: 1-10. |
| 16 | LI Z H, DU C M, ZHONG Y H, et al.. Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens [J]. Appl. Microbiol. Biotechnol., 2010, 87(3): 1065-1076. |
| 17 | XI K, SHAN L, YANG Y, et al.. Species diversity and chemotypes of Fusarium species associated with maize stalk rot in Yunnan Province of Southwest China[J/OL]. Front. Microbiol., 2021, 12: 652062[2021-10-20]. . |
| 18 | MARTINS M P, GOMES E V, SANCHES P R, et al.. Mus-52 disruption and metabolic regulation in Neurospora crassa: transcriptional responses to extracellular phosphate availability[J/OL]. PLoS ONE, 2018, 13(4): e0195871[2023-10-30]. . |
| 19 | ZHOU J, LI S M. Conversion of viridicatic acid to crustosic acid by cytochrome P450 enzyme-catalysed hydroxylation and spontaneous cyclisation[J]. Appl. Microbiol. Biotechnol., 2021, 105(24): 9181-9189. |
| 20 | FENG J, LI W, HWANG S F, et al.. Enhanced gene replacement frequency in KU70 disruption strain of Stagonospora nodorum [J]. Microbiol. Res., 2012, 167(3): 173-178. |
| 21 | HOFF B, KAMEREWERD J, SIGL C, et al.. Homologous recombination in the antibiotic producer Penicillium chrysogenum: strain DeltaPcku70 shows up-regulation of genes from the HOG pathway[J]. Appl. Microbiol. Biotechnol., 2010, 85(4): 1081-1094. |
| 22 | TAKAHASHI T, MASUDA T, KOYAMA Y. Enhanced gene targeting frequency in Ku70 and Ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae [J]. Mol. Genet. Genom., 2006, 275(5): 460-470. |
| 23 | 许铭,尹志远,高明煜,等.苹果树腐烂病菌高效基因敲除受体菌株ΔVmKu80的构建[J].西北农业学报,2016,25(2):298-305. |
| XU M, YIN Z Y, GAO M Y, et al.. Construction of enhanced gene deletion frequency recipient strain ΔVmKu80 in Valsa mali[J]. Acta Agric. Boreali Occidentalis Sin., 2016, 25(2): 298-305. | |
| 24 | QI X, SU X, GUO H, et al.. A Ku70 null mutant improves gene targeting frequency in the fungal pathogen Verticillium dahliae [J]. World J. Microbiol. Biotechnol., 2015, 31(12): 1889-1897. |
| 25 | HAARMANN T, LORENZ N, TUDZYNSKI P. Use of a nonhomologous end joining deficient strain (Deltaku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis[J]. Fungal Genet. Biol., 2008, 45(1): 35-44. |
| 26 | WANG H, PERRAULT A R, TAKEDA Y, et al.. Biochemical evidence for Ku-independent backup pathways of NHEJ[J/OL]. Nucleic Acids Res., 2020, 48(9): 5200[2023-10-20]. . |
| 27 | DAI Z, POMRANING K R, DENG S, et al.. Deletion of the KU70 homologue facilitates gene targeting in Lipomyces starkeyi strain NRRL Y-11558[J]. Curr. Genet., 2019, 65(1): 269-282. |
| [1] | 李维维,张骥,陈朝儒,王智,曲娟娟,顿宝庆,李桂英,路明. CIT2基因对突变酿酒酵母木糖利用影响的研究[J]. 生物技术进展, 2016, 6(6): 455-461. |
| [2] | 高丽,谌颜,扈廷茂,李光鹏. 肌肉生长抑制素基因在哺乳动物中的最新研究进展[J]. 生物技术进展, 2014, 4(6): 381-388. |
| [3] | 苏月焱,王旭静,王志兴. 植物选择标记基因剔除技术研究进展[J]. 生物技术进展, 2013, 3(3): 157-161. |
| [4] | 肖水华,洪钦阳,林燕珍,李今煜,. 基因敲除技术在芽胞杆菌中的应用研究[J]. 生物技术进展, 2013, 3(2): 81-84. |
| [5] | 张陈,周正富,陈明,林敏,张维. 异常球菌辐射损伤修复机制及其相关功能蛋白的研究进展[J]. 生物技术进展, 2013, 3(2): 103-108. |
| [6] | 张亚旭. DNA重组技术的研究综述[J]. 生物技术进展, 2012, 2(1): 57-63. |
| [7] | 张小娟,文莹. Ku基因在微生物中的研究进展[J]. 生物技术进展, 2011, 1(1): 26-31. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部