


生物技术进展 ›› 2022, Vol. 12 ›› Issue (5): 737-745.DOI: 10.19586/j.2095-2341.2022.0041
张晓1,2( ), 李才华1, 王婧1, 张兰兰1, 牟晓雨1, 王昕玥1, 甘刘美1, 周鹏展1, 张锐2(
), 李才华1, 王婧1, 张兰兰1, 牟晓雨1, 王昕玥1, 甘刘美1, 周鹏展1, 张锐2( )
)
收稿日期:2022-03-18
接受日期:2022-04-22
出版日期:2022-09-25
发布日期:2022-09-30
通讯作者:
张锐
作者简介:张晓 E-mail: enerrgy@126.com;
基金资助:
Xiao ZHANG1,2( ), Caihua LI1, Jing WANG1, Lanlan ZHANG1, Xiaoyu MOU1, Xinyue WANG1, Liumei GAN1, Pengzhan ZHOU1, Rui ZHANG2(
), Caihua LI1, Jing WANG1, Lanlan ZHANG1, Xiaoyu MOU1, Xinyue WANG1, Liumei GAN1, Pengzhan ZHOU1, Rui ZHANG2( )
)
Received:2022-03-18
Accepted:2022-04-22
Online:2022-09-25
Published:2022-09-30
Contact:
Rui ZHANG
摘要:
植物线粒体基因组中存在RNA编辑(C-U)现象,且有完全编辑和部分编辑之分。棉花细胞质雄性不育(cytoplasmic male sterile,CMS)系和保持系线粒体基因组中atpA基因各有两个拷贝,存在基因加倍现象,但atpA基因加倍对其RNA编辑率的影响尚不清楚。通过Southern blot、染色体步移技术确定出该基因每个拷贝具体的DNA序列,发现一个拷贝是完整的,一个拷贝是截短的。用RT-PCR、环化RT-PCR方法扩增出每个拷贝相应的cDNA,结果表明:棉花CMS系和保持系atpA基因完整拷贝和截短型拷贝都转录。完整拷贝、截短型拷贝中分别存在6、4个RNA编辑位点。完整拷贝6个RNA编辑位点在保持系中的编辑率都是100%;在CMS系中编辑率分别是100%、85%、100%、92%、100%、100%。截短型拷贝4个位点在保持系中的编辑率分别是55%、37%、55%、27%;在CMS系中编辑率分别是100%、90%、100%、100%。据此推测截短型拷贝在保持系中承受选择压较小,很可能已失去功能;而在CMS系中仍承受较大选择压,很可能具有新的功能。
中图分类号:
张晓, 李才华, 王婧, 张兰兰, 牟晓雨, 王昕玥, 甘刘美, 周鹏展, 张锐. 基因加倍对棉花线粒体atpA基因RNA编辑率的影响[J]. 生物技术进展, 2022, 12(5): 737-745.
Xiao ZHANG, Caihua LI, Jing WANG, Lanlan ZHANG, Xiaoyu MOU, Xinyue WANG, Liumei GAN, Pengzhan ZHOU, Rui ZHANG. Impact of Gene Duplication on RNA Editing Rates of Mitochondrial atpA Gene in Cotton (Gossypium hirsutum L.)[J]. Current Biotechnology, 2022, 12(5): 737-745.
| 引物名称 | 引物序列(5'→3') | 长度/nt | Tm/℃ | 用途 |
|---|---|---|---|---|
| atpAF | 5'-ATTTTCAAGTGGATGAGATCGG-3' | 22 | 59 | Southern blot |
| atpAR | 5'-GATCACAGAATCCATTGACAGC-3' | 22 | 61 | Southern blot |
| atpAIF | 5'-CTGGAATTGGCACAATATCGCGAAGTGG-3' | 28 | 65 | IPCRcRT-PCR |
| atpAIR | 5'-TCAACCATTTCCCCAGCTTGAATCTCGT-3' | 28 | 63 | IPCRcRT-PCR |
| atpA-F | 5'-TTACCAGCTCGGGGATCTAATC-3' | 20 | 60 | RT-PCR |
| 1R | 5'-CACTCGCTCGCCTTCGGGTGAGG-3' | 23 | 69 | RT-PCR |
| N2R | 5'-ATCGAGTTAGAGATCGGGTTGCAGG-3' | 25 | 64 | RT-PCR |
| S2R | 5'-ATGGAAATCCTCTTTAGCAGCCTGC-3' | 25 | 62 | RT-PCR |
表1 研究使用的引物
Table 1 Primers used in this study
| 引物名称 | 引物序列(5'→3') | 长度/nt | Tm/℃ | 用途 |
|---|---|---|---|---|
| atpAF | 5'-ATTTTCAAGTGGATGAGATCGG-3' | 22 | 59 | Southern blot |
| atpAR | 5'-GATCACAGAATCCATTGACAGC-3' | 22 | 61 | Southern blot |
| atpAIF | 5'-CTGGAATTGGCACAATATCGCGAAGTGG-3' | 28 | 65 | IPCRcRT-PCR |
| atpAIR | 5'-TCAACCATTTCCCCAGCTTGAATCTCGT-3' | 28 | 63 | IPCRcRT-PCR |
| atpA-F | 5'-TTACCAGCTCGGGGATCTAATC-3' | 20 | 60 | RT-PCR |
| 1R | 5'-CACTCGCTCGCCTTCGGGTGAGG-3' | 23 | 69 | RT-PCR |
| N2R | 5'-ATCGAGTTAGAGATCGGGTTGCAGG-3' | 25 | 64 | RT-PCR |
| S2R | 5'-ATGGAAATCCTCTTTAGCAGCCTGC-3' | 25 | 62 | RT-PCR |
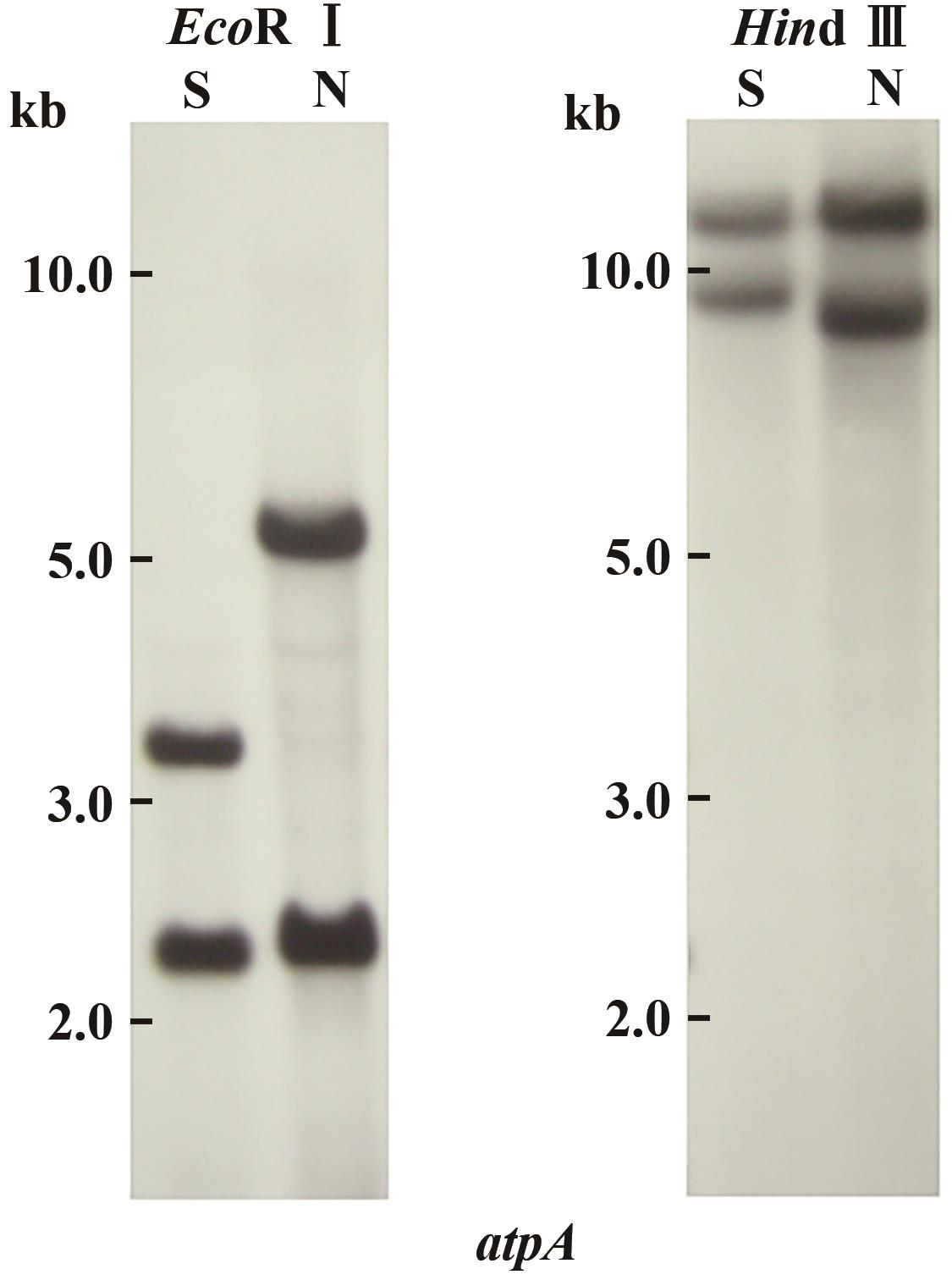
图1 陆地棉细胞质雄性不育系、保持系总DNA的Southern blot分析注:S—棉花CMS不育系;N—保持系。(N)atpA-1和(S)atpA-1含有完整的atpA基因编码序列(1 524 bp),并具有相似的3'延伸区,但是在atpA终止密码子下游161~212 bp区域,存在一个SSR位点。该位点在保持系中是(TAA)7(TA)6,在CMS中是(TAA)3(TA)2。
Fig. 1 Southern blot analysis of total DNAs of CMS, maintainer line of Gossypium hirsutum L.
| 拷贝类型 | 转录起始位点 | 转录终止位点 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 全长拷贝 | 位点/bp | -81 | -82 | -63 | -60 | -59 | +200 | +202 | +210 | ||||||
| 数目 | 10 | 3 | 2 | 1 | 1 | 10 | 6 | 1 | |||||||
| 截短拷贝 | 位点/bp | -81 | -80 | -69 | -77 | +249 | +248 | +250 | +247 | +246 | +224 | +144 | |||
| 数目 | 10 | 5 | 3 | 1 | 10 | 4 | 1 | 1 | 1 | 1 | 1 | ||||
表2 棉花CMS系全长和截短型atpA基因转录起始位点和终止位点情况
Table 2 Start and termination sites of transcripts of intact and truncated atpA gene in CMS line of cotton
| 拷贝类型 | 转录起始位点 | 转录终止位点 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 全长拷贝 | 位点/bp | -81 | -82 | -63 | -60 | -59 | +200 | +202 | +210 | ||||||
| 数目 | 10 | 3 | 2 | 1 | 1 | 10 | 6 | 1 | |||||||
| 截短拷贝 | 位点/bp | -81 | -80 | -69 | -77 | +249 | +248 | +250 | +247 | +246 | +224 | +144 | |||
| 数目 | 10 | 5 | 3 | 1 | 10 | 4 | 1 | 1 | 1 | 1 | 1 | ||||
| 编辑位点(氨基酸) | 10 | 13 | 324 | 347 | 355 | 393 | 406 | 431 | 472 | 495 | 497 | 500 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 提莫菲维小麦(T. timopheevi) | L-L | L | S-L | — | — | S-L | — | P-L | L | — | P-L | S-F |
| 普通小麦(T. aestivum) | L-L | L | S-L | — | — | S-L | — | P-L | L | — | P-L | S-F |
| 黑小麦(Triticale) | L-L | L | S-L | — | — | S-L | — | P-L | L | — | P-L | S-F |
| 月见草(Oenothera) | L | L-L | S-L | — | — | L | — | L | P-L | — | P-L | F |
| 甜菜(Beta vulgaris L.) | L | L | L | — | — | P-L | — | P-L | P-L | — | L | F |
| 棉花(Gossypium hirsutum L.) | L | L | L | P-S | S-L | L | L-F | P-L | P-L | P-L | T | K |
表3 棉花atpA编辑位点与其他植物的比较
Table 3 Comparison of cotton atpA editing sites with those of other plants
| 编辑位点(氨基酸) | 10 | 13 | 324 | 347 | 355 | 393 | 406 | 431 | 472 | 495 | 497 | 500 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 提莫菲维小麦(T. timopheevi) | L-L | L | S-L | — | — | S-L | — | P-L | L | — | P-L | S-F |
| 普通小麦(T. aestivum) | L-L | L | S-L | — | — | S-L | — | P-L | L | — | P-L | S-F |
| 黑小麦(Triticale) | L-L | L | S-L | — | — | S-L | — | P-L | L | — | P-L | S-F |
| 月见草(Oenothera) | L | L-L | S-L | — | — | L | — | L | P-L | — | P-L | F |
| 甜菜(Beta vulgaris L.) | L | L | L | — | — | P-L | — | P-L | P-L | — | L | F |
| 棉花(Gossypium hirsutum L.) | L | L | L | P-S | S-L | L | L-F | P-L | P-L | P-L | T | K |
| 编辑位点 | 1 039 bp | 1 064 bp | 1 216 bp | 1 292 bp | 1 415 bp | 1 484 bp |
|---|---|---|---|---|---|---|
| 氨基酸的变化 | P-S | S-L | L-F | P-L | P-L | P-L |
| (N)atpA-1 | 100% | 100% | 100% | 100% | 100% | 100% |
| (S)atpA-1 | 100% | 85% | 100% | 92% | 100% | 100% |
| (N)atpA-2 | 55% | 37% | 55% | 27% | — | — |
| (S)atpA-2 | 100% | 90% | 100% | 100% | — | — |
表4 棉花CMS系(S)和保持系(N)atpA基因完整拷贝和截短型拷贝的RNA编辑率比较
Table 4 Comparison of RNA editing rates of intact and truncated copies of atpA gene in CMS line(S) and maintainer line(N) of cotton
| 编辑位点 | 1 039 bp | 1 064 bp | 1 216 bp | 1 292 bp | 1 415 bp | 1 484 bp |
|---|---|---|---|---|---|---|
| 氨基酸的变化 | P-S | S-L | L-F | P-L | P-L | P-L |
| (N)atpA-1 | 100% | 100% | 100% | 100% | 100% | 100% |
| (S)atpA-1 | 100% | 85% | 100% | 92% | 100% | 100% |
| (N)atpA-2 | 55% | 37% | 55% | 27% | — | — |
| (S)atpA-2 | 100% | 90% | 100% | 100% | — | — |
| 1 | 张晓,张锐,孙国清,等.优化的反向PCR结合TAIL-PCR法克隆棉花线粒体atpA双拷贝基因及其侧翼序列[J].生物工程学报,2012,28(1): 104-115. |
| 2 | CHATEIGNER-BOUTIN A L, SMALL I. Plant RNA editing[J]. RNA Biol., 2014, 7(2): 213-219. |
| 3 | 张晓,张锐,侯思宇,等.高等植物线粒体基因组研究进展[J].中国农业科技导报,2011, 13(4): 23-31. |
| 4 | SATOH M, KUBO T, MIKAMI T.The Owen mitochondrial genome in sugar beet (Beta vulgaris L.): possible mechanisms of extensive rearrangements and the origin of the mitotype-unique regions[J]. Theor. Appl. Genet.,2006, 113(3): 477-484. |
| 5 | HANDA H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana [J]. Nucl. Acids Res., 2003,31(20): 5907-5916. |
| 6 | PICARDI E, HORNER D S, CHIARA M,et al.. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing[J]. Nucl. Acids Res., 2010,38(14):4755-4767. |
| 7 | NOTSU Y, MASOOD S, NISHIKAWA T, et al.. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants[J]. Mol. Genet. Genom., 2002, 268(4): 434-445. |
| 8 | GIEGÉ P, BRENNICKE A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs[J]. Proc. Natl. Acad. Sci. USA, 1999,96(26):15324-15329. |
| 9 | FANG Y, WU H, ZHANG T, et al.. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome[J/OL]. PLoS ONE, 2012, 7(5): e37164[2022-04-19]. . |
| 10 | HE P, XIAO G, LIU H, et al.. Two pivotal RNA editing sites in the mitochondrial atp1mRNA are required for ATP synthase to produce sufficient ATP for cotton fiber cell elongation[J]. New Phytol., 2018, 218(1): 167-182. |
| 11 | CHAW S V M, SHIH A V C, WANG D, et al.. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites[J]. Mol. Biol. Evol., 2008, 25(3): 603-615. |
| 12 | SMALL I D, PEETERS N. The PPR motif-a TPR-related motif prevalent in plant organellar proteins[J]. Trends Biochem. Sci., 2000, 25(2): 46-47. |
| 13 | 何鹏,陈海燕,俞嘉宁.PPR蛋白参与RNA编辑机制的研究进展[J].西北植物学报,2013,33(2):415-421. |
| 14 | 陈倩,刘石锋,洪广成,等.植物线粒体RNA编辑调控研究进展[J].分子植物育种,2019,17(3):869-876. |
| 15 | DONIWA Y, UETA M, MASAMI U,et al.. The involvement of a PPR protein of the P subfamily in partial RNA editing of an Arabidopsis mitochondrial transcript[J]. Gene, 2010, 454 (1-2): 39-46. |
| 16 | OKUDA K, HABATA Y, KOBAYASHI Y, et al.. Amino acid sequence variations in Nicotiana CRR4 orthologs determine the species specific efficiency of RNA editing in plastids[J]. Nucl. Acid. Res., 2008, 36(19): 6155-6164. |
| 17 | MYOUGA, OKUDA K, MOTOHASHI R, et al.. Conserved domain structure of pentatricopeptide repeat protein involved in chloroplast RNA editing[J]. Proc. Natl. Acad. Sci. USA, 2007, 104(19): 8178-8183. |
| 18 | CHATEIGNER B A L, RAMOS-VEGA M, GUEVARA-GARCÍA A, et al.. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts[J]. Plant J., 2008, 56(4): 590-602. |
| 19 | SUNG T Y, TSENG C C, HSIEH M H. The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria [J]. Plant J., 2010, 63(3): 499-511. |
| 20 | ZHU Q, DUGARDEYN J, ZHANG C, et al.. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism[J]. Plant J., 2010,71(5): 836-849. |
| 21 | VERBITSKIY D, MERWE J A, ZEHRMANN A, et al.. The E-Class PPR protein MEF3 of Arabidopsis thaliana can also function in mitochondrial RNA editing with an additional DYW domain[J]. Plant Cell Phys., 2011, 53(2): 358-367. |
| 22 | OKUDA K, CHATEIGNER-BOUTIN A L, NAKAMURA T, et al.. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts [J]. Plant Cell, 2009, 21(1):146-156. |
| 23 | ZHOU W, CHENG Y, YAP A, et al.. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth[J]. Plant J., 2009, 58(1): 82-96. |
| 24 | CAO Z L, YU Q B, SUN Y, et al.. A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development[J]. J. Integr. Plant Biol., 2011, 53(4): 258-269. |
| 25 | YU Q B, JIANG Y, CHONG K, et al.. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana [J]. Plant J., 2009,59(6): 1011-1023. |
| 26 | CAI W, JI D, PENG L, et al.. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis [J]. Plant Physl., 2009, 150(3): 1260-1271. |
| 27 | ROBBINS J C, HELLER W P, HANSON M R. A comparative genomics approach identifies a PPR-DYW protein that is essential for C to U editing of the Arabidopsis chloroplast accD transcript[J]. RNA,2009,15(6): 1142-1153. |
| 28 | HAMMANI K, OKUDA K, TANZ-SANDRA K, et al.. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites[J]. Plant Cell, 2009, 21(11): 3686-3699. |
| 29 | HAMMANI K, FRANCS-SMALL C C, TAKENAKA M, et al.. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria[J]. J. Biol. Chem., 2011, 286(24): 21361-21371. |
| 30 | ZEHRMANN A, VERNITSKIY D, VANDEMERWE J A, et al.. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana [J]. Plant Cell, 2009, 21(2): 558-567. |
| 31 | ZEHRMANN A, MERWE JVAN DER, VERBITSKIY D, et al.. The DYW-class PPR protein MEF7 is required for RNA editing at four sites in mitochondria of Arabidopsis Thaliana [J]. RNA Biol., 2012, 9 (2): 155-161. |
| 32 | TAKENAKA M. MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis [J]. Plant Phys., 2012,152 (2):939-947. |
| 33 | VERBITSKIY D, ZEHRMANN A, BRENNICKE A, et al.. A truncated MEF11 protein shows site-specific effects on mitochondrial RNA editing[J]. Plant Sign. Beh., 2010, 5(5): 558-560. |
| 34 | VERBITSKIY D, HÄRTEL B, ZEHRMANN A, et al.. The DYW-E-PPR protein MEF14 is required for RNA editing at site matR-1895 in mitochondria of Arabidopsis thaliana [J]. Sci Dir., 2011, 585(4): 700-704. |
| 35 | SALONE V, RUDINGER M, PLOSAKIEWICZ M, et al.. A hypothesis on the identification of the editing enzyme in plant organelles[J]. FEBS Lett., 2007, 581(22): 4132-4138. |
| 36 | 马艳莉,俞嘉宁.高等植物叶绿体RNA编辑研究进展[J].生命科学,2009,21(3):439-443. |
| 37 | ZHANG J. Evolution by gene duplication: an update[J]. Trends Ecol Evol., 2003, 18(6): 292-298. |
| 38 | 孙红正,葛颂.重复基因的进化—回顾与进展[J].植物学报,2010,45(1):13-22. |
| 39 | OHNO S. Evolution by Gene Duplication[M]. Berlin: Springer-Verlag, 1970. |
| 40 | OGIHARA Y, YAMAZAKI Y, MURAI K, et al.. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome[J]. Nucl. Acids Res., 2005, 33(19): 6235-6250. |
| 41 | CLIFTON S W, MINX P, FAURON C M, et al.. Sequence and comparative analysis of the maize NB mitochondrial genome[J]. Plant Phys., 2004, 136(3): 3486-3503. |
| 42 | UNSELD M, MARIENFELD J R, BRANDT P, et al.. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides[J]. Nat. Genet., 1997, 15(1): 57-61. |
| 43 | KUBO T, NISHIZAWA S, SUGAWARA A, et al.. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys (GCA)[J]. Nucl. Acids Res., 2000, 28(13): 2571-2576. |
| 44 | HANDA H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana [J]. Nucl. Acids Res., 2003, 31(20): 5907-5916. |
| 45 | ALVERSON A J, WEI X X, RICE D W, et al.. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae)[J]. Mol. Biol. Evol., 2010, 27(6): 1436-1448. |
| 46 | 张晓,孟志刚,孙国清,等.陆地棉线粒体nad6基因mRNA无常规终止密码子[J]. 中国生物化学与分子生物学报,2014,30(11):1119-1125. |
| 47 | 张晓,张锐, 史计,等.陆地棉胞质雄性不育系与保持系线粒体基因组RFLP分析[J].中国农业科学,2012,45(2):208-217. |
| 48 | KUHN J, BINDER S. RT-PCR analysis of 5' to 3'-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria[J]. Nucl. Acids Res., 2002, 30(2): 439-446. |
| 49 | KUREK I, EZRA D, BEGU D, et al.. Studies on the effects of nuclear background and tissue specificity on RNA editing of the mitochondrial ATP synthase subunits α, 6 and 9 in fertile and cytoplasmic male-sterile (CMS) wheat[J]. Theor. Appl. Genet., 1997, 95(8): 1305-1311. |
| 50 | LASER B, KÜCK U. The mitochondrial atpA/atp9 co-transcript in wheat and triticale: RNA processing depends on the nuclear genotype[J]. Curr. Genet., 1995, 29(1): 50-57. |
| 51 | SCHUSTER W, BRENNICKE A. RNA editing of ATPase subunit 9 transcripts in Oenothera mitochondria [J]. FEBS Lett., 1990, 268(1): 252-256. |
| 52 | SENDA M, ONODERA Y, MIKAMI T. Recombination events across the atpA-associated repeated sequences in the mitochondrial genomes of beets[J]. Theor. Appl. Genet., 1998, 96(6-7): 964-968. |
| 53 | OGIHARA Y, YAMAZAKI Y, MURAI K, et al.. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome[J]. Nucl. Acids Res., 2005, 33(19): 6235-6250. |
| 54 | CHEN Z, NIE H, GROVER C E, et al.. Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species[J]. Plant Biol., 2017, 19(3): 484-493. |
| 55 | TANG M, CHEN Z, GROVER C E, et al.. Rapid evolutionary divergence of Gossypium barbadense and G. hirsutum mitochondrial genomes[J]. BMC Genom., 2015, 16(1): 770. |
| 56 | 雷彬彬,李双双,刘国政,等.高等植物线粒体基因组进化分析[J].分子植物育种,2012,10(4):490-500. |
| 57 | IWAHASHI M, NAKAZONO M, KANNO A, et al.. Genetic and physical maps and a clone bank of mitochondrial DNA from rice[J]. Theor. Appl. Genet., 1992, 84(3-4): 275-279. |
| 58 | CLIFTON S W, MINX P, FAURON C M, et al.. Sequence and comparative analysis of the maize NB mitochondrial genome[J]. Plant Phys., 2004, 136(3): 3486-3503. |
| 59 | LNSDALE D M, BREARS T, HODGE T P, et al.. The plant mitochondrial genome: homologous recombination as a mechanism for generating heterogeneity[J]. Phil. Trans. R Soc. Lond B, 1988, 319(1193): 149-163. |
| 60 | MARECHAL A, BRISSON N. Recombination and the maintenance of plant organelle genome stability[J]. New Phytol., 2010, 186(2): 299-317. |
| 61 | WATERS E R, NGUYEN S L, ESKANDAR R, et al.. The recent evolution of a pseudogene: diversity and divergence of a mitochondria-localized small heat shock protein in Arabidopsis thaliana [J]. Genome, 2008, 51(3): 177-186. |
| 62 | MÜLLER K, STORCHOVA H. Transcription of atp1 is influenced by both genomic configuration and nuclear background in the highly rearranged mitochondrial genomes of Silene vulgaris [J]. Plant Mol Biol., 2013, 81(4-5): 495-505. |
| [1] | 肖荣, 魏云晓, 王远, 孟志刚, 梁成真, 陈全家, 张锐. 茎尖法转基因棉花植株真实性鉴定方法探究[J]. 生物技术进展, 2022, 12(1): 83-89. |
| [2] | 高正银,孙文杰,宋晓云,胡轼,左开井. 雷蒙德棉第Ⅲ类过氧化物酶全基因组鉴定和表达分析[J]. 生物技术进展, 2019, 9(5): 490-501. |
| [3] | 王裴林,周利利,梁成真,孟志刚,郭三堆,张锐. 棉花线粒体基因cRT-PCR改良及其在寻找CMS相关基因中的应用[J]. 生物技术进展, 2019, 9(3): 303-308. |
| [4] | 胡文冉,李晓东,周小云,李晓荣,杨洋,李波. 棉花GhCAD6基因表达及其功能分析研究[J]. 生物技术进展, 2019, 9(1): 46-53. |
| [5] | 宋志红,孟庆忠,张涛,张胜昔,王贵春,李国荣. ISSR分子标记在棉花遗传育种上的应用[J]. 生物技术进展, 2014, 4(6): 411-414. |
| [6] | 杨淑巧,王志安,张安红,许琦,肖娟丽,罗晓丽. 棉花WRKY基因GhWRKY25的克隆和表达分析[J]. 生物技术进展, 2014, 4(4): 274-279. |
| [7] | 石雅丽,张锐,任茂智,孟志刚,周焘,孙国清,孟钊红,郭三堆. 棉花雄性不育的研究进展[J]. 生物技术进展, 2013, 3(5): 328-335. |
| [8] | 马维军,任爱民,尹国,张玉娟,韩秋成,崔明晖. 利用多重PCR-SSR标记鉴别三系杂交棉种子混杂和SSR位点杂合[J]. 生物技术进展, 2013, 3(4): 248-251. |
| [9] | 罗晓丽,王志安,肖娟丽,张安红,吴家和. 不同继代时间调控棉花愈伤组织褐化死亡和植株再生效率的研究[J]. 生物技术进展, 2013, 3(3): 201-205. |
| [10] | 孙国清,张锐,王远,任茂智,孟志刚,周焘,陈全家,曲延英,郭三堆. 转基因抗虫棉抗性纯度对产量的影响[J]. 生物技术进展, 2013, 3(1): 27-31. |
| [11] | 石雅丽,张锐,孟志刚,郭三堆. 一种高效提取棉花RNA的改良方法[J]. 生物技术进展, 2011, 1(3): 219-222. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2021《生物技术进展》编辑部